Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Array image and a genomic representation of a normalized arrayCGHDescriptionDisplays an array image and a genomic representation of a normalized arrayCGH. Usage
## S3 method for class 'arrayCGH'
report.plot(arrayCGH, x="PosOrder", y=c("LogRatioNorm",
"LogRatio"), chrLim=NULL, layout=TRUE, main=NULL, zlim=NULL, ...)
## Default S3 method:
report.plot(spot.data, clone.data, design, x="PosOrder",
y=c("LogRatioNorm", "LogRatio"), chrLim=NULL, layout=TRUE, main=NULL,
zlim=NULL, ...)
Arguments
DetailsThis function successively calls NotePeople interested in tools for array-CGH analysis can visit our web-page: http://bioinfo.curie.fr. Author(s)Pierre Neuvial, manor@curie.fr. See Also
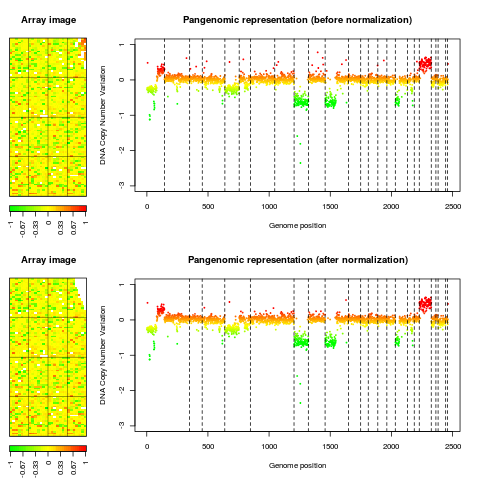
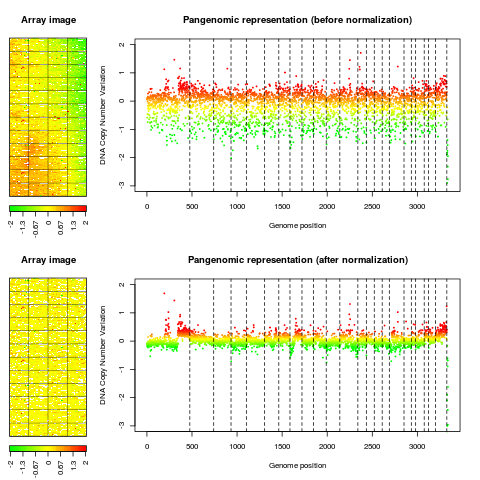
Examplesdata(spatial) ### edge: local spatial bias ## aggregate arrayCGH without normalization for comparison with ## normalized array edge.nonorm <- norm(edge, flag.list=NULL, FUN=median, na.rm=TRUE) edge.nonorm <- sort(edge.nonorm, position.var="PosOrder") layout(matrix(c(1,2,4,5,3,3,6,6), 4,2),width=c(1, 4), height=c(6,1,6,1)) report.plot(edge.nonorm, chrLim="LimitChr", layout=FALSE, main="Pangenomic representation (before normalization)", zlim=c(-1,1), ylim=c(-3,1)) report.plot(edge.norm, chrLim="LimitChr", layout=FALSE, main="Pangenomic representation (after normalization)", zlim=c(-1,1), ylim=c(-3,1)) ### gradient: global array Trend ## aggregate arrayCGH without normalization for comparison with ## normalized array gradient.nonorm <- norm(gradient, flag.list=NULL, FUN=median, na.rm=TRUE) gradient.nonorm <- sort(gradient.nonorm) layout(matrix(c(1,2,4,5,3,3,6,6), 4,2),width=c(1, 4), height=c(6,1,6,1)) report.plot(gradient.nonorm, chrLim="LimitChr", layout=FALSE, main="Pangenomic representation (before normalization)", zlim=c(-2,2), ylim=c(-3,2)) report.plot(gradient.norm, chrLim="LimitChr", layout=FALSE, main="Pangenomic representation (after normalization)", zlim=c(-2,2), ylim=c(-3,2)) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(MANOR)
Loading required package: GLAD
######################################################################################
Have fun with GLAD
For smoothing it is possible to use either
the AWS algorithm (Polzehl and Spokoiny, 2002,
or the HaarSeg algorithm (Ben-Yaacov and Eldar, Bioinformatics, 2008,
If you use the package with AWS, please cite:
Hupe et al. (Bioinformatics, 2004, and Polzehl and Spokoiny (2002,
If you use the package with HaarSeg, please cite:
Hupe et al. (Bioinformatics, 2004, and (Ben-Yaacov and Eldar, Bioinformatics, 2008,
For fast computation it is recommanded to use
the daglad function with smoothfunc=haarseg
######################################################################################
New options are available in daglad: see help for details.
Attaching package: 'MANOR'
The following object is masked from 'package:base':
norm
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/MANOR/report.plot.Rd_%03d_medium.png", width=480, height=480)
> ### Name: report.plot
> ### Title: Array image and a genomic representation of a normalized
> ### arrayCGH
> ### Aliases: report.plot report.plot.arrayCGH report.plot.default
> ### Keywords: hplot
>
> ### ** Examples
>
> data(spatial)
>
> ### edge: local spatial bias
> ## aggregate arrayCGH without normalization for comparison with
> ## normalized array
> edge.nonorm <- norm(edge, flag.list=NULL, FUN=median, na.rm=TRUE)
> edge.nonorm <- sort(edge.nonorm, position.var="PosOrder")
>
> layout(matrix(c(1,2,4,5,3,3,6,6), 4,2),width=c(1, 4), height=c(6,1,6,1))
> report.plot(edge.nonorm, chrLim="LimitChr", layout=FALSE,
+ main="Pangenomic representation (before normalization)", zlim=c(-1,1),
+ ylim=c(-3,1))
> report.plot(edge.norm, chrLim="LimitChr", layout=FALSE,
+ main="Pangenomic representation (after normalization)", zlim=c(-1,1),
+ ylim=c(-3,1))
>
> ### gradient: global array Trend
> ## aggregate arrayCGH without normalization for comparison with
> ## normalized array
> gradient.nonorm <- norm(gradient, flag.list=NULL, FUN=median, na.rm=TRUE)
> gradient.nonorm <- sort(gradient.nonorm)
>
> layout(matrix(c(1,2,4,5,3,3,6,6), 4,2),width=c(1, 4), height=c(6,1,6,1))
> report.plot(gradient.nonorm, chrLim="LimitChr", layout=FALSE,
+ main="Pangenomic representation (before normalization)", zlim=c(-2,2),
+ ylim=c(-3,2))
> report.plot(gradient.norm, chrLim="LimitChr", layout=FALSE,
+ main="Pangenomic representation (after normalization)", zlim=c(-2,2),
+ ylim=c(-3,2))
>
>
>
>
>
> dev.off()
null device
1
>
|