Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Calculate Double Principle Coordinate Analysis (DPCoA) using phylogenetic distanceDescriptionFunction uses abundance ( UsageDPCoA(physeq, correction = cailliez, scannf = FALSE, ...) Arguments
DetailsIn most real-life, real-data applications, the phylogenetic tree
will not provide a Euclidean distance matrix, and so a correction
will be performed, if needed. See ValueA Author(s)Julia Fukuyama julia.fukuyama@gmail.com. Adapted for phyloseq by Paul J. McMurdie. ReferencesPavoine, S., Dufour, A.B. and Chessel, D. (2004) From dissimilarities among species to dissimilarities among communities: a double principal coordinate analysis. Journal of Theoretical Biology, 228, 523-537. See Also
Examples# # # # # # Esophagus data(esophagus) eso.dpcoa <- DPCoA(esophagus) eso.dpcoa plot_ordination(esophagus, eso.dpcoa, "samples") plot_ordination(esophagus, eso.dpcoa, "species") plot_ordination(esophagus, eso.dpcoa, "biplot") # # # # # # # # GlobalPatterns data(GlobalPatterns) # subset GP to top-150 taxa (to save computation time in example) keepTaxa <- names(sort(taxa_sums(GlobalPatterns), TRUE)[1:150]) GP <- prune_taxa(keepTaxa, GlobalPatterns) # Perform DPCoA GP.dpcoa <- DPCoA(GP) plot_ordination(GP, GP.dpcoa, color="SampleType") Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(phyloseq)
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/phyloseq/DPCoA.Rd_%03d_medium.png", width=480, height=480)
> ### Name: DPCoA
> ### Title: Calculate Double Principle Coordinate Analysis (DPCoA) using
> ### phylogenetic distance
> ### Aliases: DPCoA
>
> ### ** Examples
>
> # # # # # # Esophagus
> data(esophagus)
> eso.dpcoa <- DPCoA(esophagus)
> eso.dpcoa
Double principal coordinate analysis
call: dpcoa(df = data.frame(OTU), dis = patristicDist, scannf = scannf)
class: dpcoa
$nf (axis saved) : 2
$rank: 2
eigen values: 0.001447 0.0003375
vector length mode content
1 $dw 58 numeric category weights
2 $lw 3 numeric collection weights
3 $eig 2 numeric eigen values
data.frame nrow ncol content
1 $dls 58 2 coordinates of the categories
2 $li 3 2 coordinates of the collections
3 $c1 57 2 scores of the principal axes of the categories
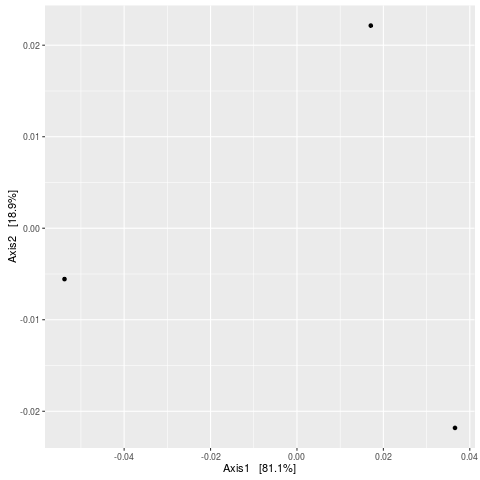
> plot_ordination(esophagus, eso.dpcoa, "samples")
No available covariate data to map on the points for this plot `type`
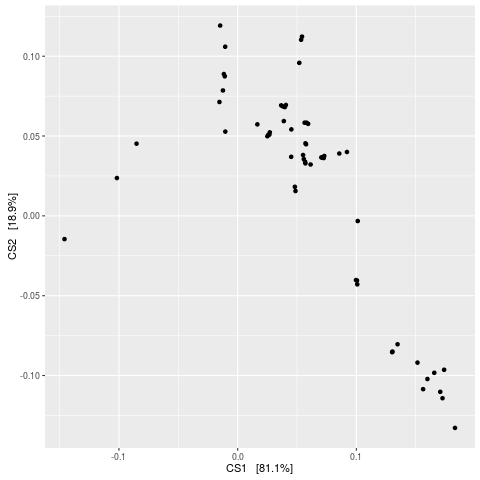
> plot_ordination(esophagus, eso.dpcoa, "species")
No available covariate data to map on the points for this plot `type`
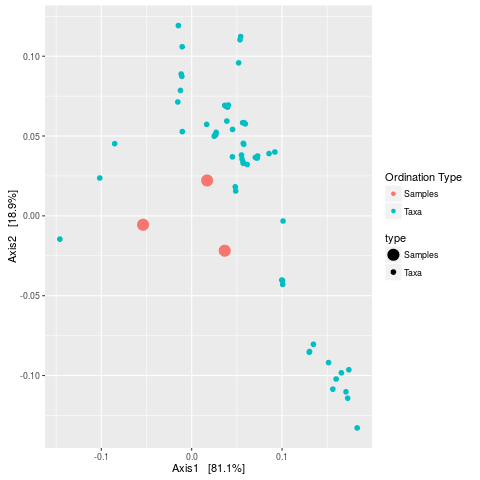
> plot_ordination(esophagus, eso.dpcoa, "biplot")
> #
> #
> # # # # # # GlobalPatterns
> data(GlobalPatterns)
> # subset GP to top-150 taxa (to save computation time in example)
> keepTaxa <- names(sort(taxa_sums(GlobalPatterns), TRUE)[1:150])
> GP <- prune_taxa(keepTaxa, GlobalPatterns)
> # Perform DPCoA
> GP.dpcoa <- DPCoA(GP)
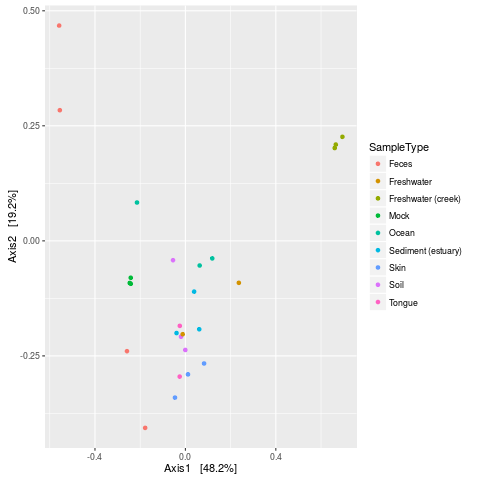
> plot_ordination(GP, GP.dpcoa, color="SampleType")
>
>
>
>
>
> dev.off()
null device
1
>
|