Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Class "ChromosomeModels"DescriptionCollection of dependency models fitting two data sets in particular chromosome. Objects from the Class Function
Slots
Methods
. Author(s)Olli-Pekka Huovilainen ohuovila@gmail.com See Also For calculation of dependency models for chromosomal arm:
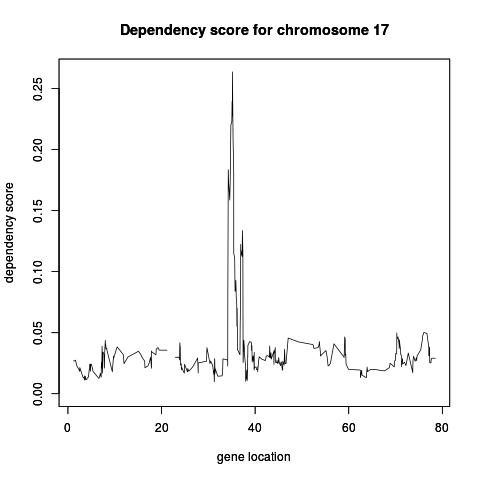
Examplesdata(chromosome17) ## calculate dependency models over chromosome 17 model17 <- screen.cgh.mrna(geneExp, geneCopyNum, windowSize = 10, chr = 17) model17 ## Information of the dependency model which has the highest dependency score topGenes(model17, 1) ## Finding a dependency model by its name findModel(model17, "ENSG00000129250") ## Information of the first dependency model model17[[1]] #Plotting plot(model17) # genes in p arm with the highest dependency scores topGenes(model17[['p']], 5) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(pint)
Loading required package: mvtnorm
Loading required package: Matrix
Loading required package: dmt
Loading required package: MASS
dmt Copyright (C) 2008-2013 Leo Lahti and Olli-Pekka Huovilainen.
This program comes with ABSOLUTELY NO
WARRANTY.
This is free software, and you are welcome to redistribute it
under the FreeBSD license.
pint Copyright (C) 2008-2013 Olli-Pekka Huovilainen and Leo Lahti.
This program comes with ABSOLUTELY NO WARRANTY.
This is free software, and you are welcome to redistribute it
under the FreeBSD license.
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/pint/ChromosomeModels-class.Rd_%03d_medium.png", width=480, height=480)
> ### Name: ChromosomeModels-class
> ### Title: Class "ChromosomeModels"
> ### Aliases: ChromosomeModels-class getPArm getQArm [[ [[<- getChromosome
> ### getArm getGeneName getModelMethod getParams getWindowSize topGenes
> ### topModels getModelNumbers isEmpty orderGenes findModel
> ### [[,ChromosomeModels-method [[<-,ChromosomeModels-method
> ### getChromosome,ChromosomeModels-method getPArm,ChromosomeModels-method
> ### getQArm,ChromosomeModels-method getArm,ChromosomeModels-method
> ### getGeneName,ChromosomeModels-method getLoc,ChromosomeModels-method
> ### getScore,ChromosomeModels-method
> ### getModelMethod,ChromosomeModels-method
> ### getParams,ChromosomeModels-method
> ### getWindowSize,ChromosomeModels-method
> ### topGenes,ChromosomeModels-method topModels,ChromosomeModels-method
> ### getModelNumbers,ChromosomeModels-method
> ### isEmpty,ChromosomeModels-method orderGenes,ChromosomeModels-method
> ### findModel,ChromosomeModels-method isEmpty,ChromosomeModels-method
> ### [[,ChromosomeModels-method as.data.frame,ChromosomeModels-method
> ### Keywords: classes
>
> ### ** Examples
>
> data(chromosome17)
>
> ## calculate dependency models over chromosome 17
> model17 <- screen.cgh.mrna(geneExp, geneCopyNum, windowSize = 10, chr
+ = 17)
Imputing missing values..
Imputing missing values..
Matching probes between the data sets..
Calculating dependency models for 17p with method pSimCCA, window size:10
17p; window 1/92
17p; window 2/92
17p; window 3/92
17p; window 4/92
17p; window 5/92
17p; window 6/92
17p; window 7/92
17p; window 8/92
17p; window 9/92
17p; window 10/92
17p; window 11/92
17p; window 12/92
17p; window 13/92
17p; window 14/92
17p; window 15/92
17p; window 16/92
17p; window 17/92
17p; window 18/92
17p; window 19/92
17p; window 20/92
17p; window 21/92
17p; window 22/92
17p; window 23/92
17p; window 24/92
17p; window 25/92
17p; window 26/92
17p; window 27/92
17p; window 28/92
17p; window 29/92
17p; window 30/92
17p; window 31/92
17p; window 32/92
17p; window 33/92
17p; window 34/92
17p; window 35/92
17p; window 36/92
17p; window 37/92
17p; window 38/92
17p; window 39/92
17p; window 40/92
17p; window 41/92
17p; window 42/92
17p; window 43/92
17p; window 44/92
17p; window 45/92
17p; window 46/92
17p; window 47/92
17p; window 48/92
17p; window 49/92
17p; window 50/92
17p; window 51/92
17p; window 52/92
17p; window 53/92
17p; window 54/92
17p; window 55/92
17p; window 56/92
17p; window 57/92
17p; window 58/92
17p; window 59/92
17p; window 60/92
17p; window 61/92
17p; window 62/92
17p; window 63/92
17p; window 64/92
17p; window 65/92
17p; window 66/92
17p; window 67/92
17p; window 68/92
17p; window 69/92
17p; window 70/92
17p; window 71/92
17p; window 72/92
17p; window 73/92
17p; window 74/92
17p; window 75/92
17p; window 76/92
17p; window 77/92
17p; window 78/92
17p; window 79/92
17p; window 80/92
17p; window 81/92
17p; window 82/92
17p; window 83/92
17p; window 84/92
17p; window 85/92
17p; window 86/92
17p; window 87/92
17p; window 88/92
17p; window 89/92
17p; window 90/92
17p; window 91/92
17p; window 92/92
Calculating dependency models for 17q with method pSimCCA, window size:10
17q; window 1/274
17q; window 2/274
17q; window 3/274
17q; window 4/274
17q; window 5/274
17q; window 6/274
17q; window 7/274
17q; window 8/274
17q; window 9/274
17q; window 10/274
17q; window 11/274
17q; window 12/274
17q; window 13/274
17q; window 14/274
17q; window 15/274
17q; window 16/274
17q; window 17/274
17q; window 18/274
17q; window 19/274
17q; window 20/274
17q; window 21/274
17q; window 22/274
17q; window 23/274
17q; window 24/274
17q; window 25/274
17q; window 26/274
17q; window 27/274
17q; window 28/274
17q; window 29/274
17q; window 30/274
17q; window 31/274
17q; window 32/274
17q; window 33/274
17q; window 34/274
17q; window 35/274
17q; window 36/274
17q; window 37/274
17q; window 38/274
17q; window 39/274
17q; window 40/274
17q; window 41/274
17q; window 42/274
17q; window 43/274
17q; window 44/274
17q; window 45/274
17q; window 46/274
17q; window 47/274
17q; window 48/274
17q; window 49/274
17q; window 50/274
17q; window 51/274
17q; window 52/274
17q; window 53/274
17q; window 54/274
17q; window 55/274
17q; window 56/274
17q; window 57/274
17q; window 58/274
17q; window 59/274
17q; window 60/274
17q; window 61/274
17q; window 62/274
17q; window 63/274
17q; window 64/274
17q; window 65/274
17q; window 66/274
17q; window 67/274
17q; window 68/274
17q; window 69/274
17q; window 70/274
17q; window 71/274
17q; window 72/274
17q; window 73/274
17q; window 74/274
17q; window 75/274
17q; window 76/274
17q; window 77/274
17q; window 78/274
17q; window 79/274
17q; window 80/274
17q; window 81/274
17q; window 82/274
17q; window 83/274
17q; window 84/274
17q; window 85/274
17q; window 86/274
17q; window 87/274
17q; window 88/274
17q; window 89/274
17q; window 90/274
17q; window 91/274
17q; window 92/274
17q; window 93/274
17q; window 94/274
17q; window 95/274
17q; window 96/274
17q; window 97/274
17q; window 98/274
17q; window 99/274
17q; window 100/274
17q; window 101/274
17q; window 102/274
17q; window 103/274
17q; window 104/274
17q; window 105/274
17q; window 106/274
17q; window 107/274
17q; window 108/274
17q; window 109/274
17q; window 110/274
17q; window 111/274
17q; window 112/274
17q; window 113/274
17q; window 114/274
17q; window 115/274
17q; window 116/274
17q; window 117/274
17q; window 118/274
17q; window 119/274
17q; window 120/274
17q; window 121/274
17q; window 122/274
17q; window 123/274
17q; window 124/274
17q; window 125/274
17q; window 126/274
17q; window 127/274
17q; window 128/274
17q; window 129/274
17q; window 130/274
17q; window 131/274
17q; window 132/274
17q; window 133/274
17q; window 134/274
17q; window 135/274
17q; window 136/274
17q; window 137/274
17q; window 138/274
17q; window 139/274
17q; window 140/274
17q; window 141/274
17q; window 142/274
17q; window 143/274
17q; window 144/274
17q; window 145/274
17q; window 146/274
17q; window 147/274
17q; window 148/274
17q; window 149/274
17q; window 150/274
17q; window 151/274
17q; window 152/274
17q; window 153/274
17q; window 154/274
17q; window 155/274
17q; window 156/274
17q; window 157/274
17q; window 158/274
17q; window 159/274
17q; window 160/274
17q; window 161/274
17q; window 162/274
17q; window 163/274
17q; window 164/274
17q; window 165/274
17q; window 166/274
17q; window 167/274
17q; window 168/274
17q; window 169/274
17q; window 170/274
17q; window 171/274
17q; window 172/274
17q; window 173/274
17q; window 174/274
17q; window 175/274
17q; window 176/274
17q; window 177/274
17q; window 178/274
17q; window 179/274
17q; window 180/274
17q; window 181/274
17q; window 182/274
17q; window 183/274
17q; window 184/274
17q; window 185/274
17q; window 186/274
17q; window 187/274
17q; window 188/274
17q; window 189/274
17q; window 190/274
17q; window 191/274
17q; window 192/274
17q; window 193/274
17q; window 194/274
17q; window 195/274
17q; window 196/274
17q; window 197/274
17q; window 198/274
17q; window 199/274
17q; window 200/274
17q; window 201/274
17q; window 202/274
17q; window 203/274
17q; window 204/274
17q; window 205/274
17q; window 206/274
17q; window 207/274
17q; window 208/274
17q; window 209/274
17q; window 210/274
17q; window 211/274
17q; window 212/274
17q; window 213/274
17q; window 214/274
17q; window 215/274
17q; window 216/274
17q; window 217/274
17q; window 218/274
17q; window 219/274
17q; window 220/274
17q; window 221/274
17q; window 222/274
17q; window 223/274
17q; window 224/274
17q; window 225/274
17q; window 226/274
17q; window 227/274
17q; window 228/274
17q; window 229/274
17q; window 230/274
17q; window 231/274
17q; window 232/274
17q; window 233/274
17q; window 234/274
17q; window 235/274
17q; window 236/274
17q; window 237/274
17q; window 238/274
17q; window 239/274
17q; window 240/274
17q; window 241/274
17q; window 242/274
17q; window 243/274
17q; window 244/274
17q; window 245/274
17q; window 246/274
17q; window 247/274
17q; window 248/274
17q; window 249/274
17q; window 250/274
17q; window 251/274
17q; window 252/274
17q; window 253/274
17q; window 254/274
17q; window 255/274
17q; window 256/274
17q; window 257/274
17q; window 258/274
17q; window 259/274
17q; window 260/274
17q; window 261/274
17q; window 262/274
17q; window 263/274
17q; window 264/274
17q; window 265/274
17q; window 266/274
17q; window 267/274
17q; window 268/274
17q; window 269/274
17q; window 270/274
17q; window 271/274
17q; window 272/274
17q; window 273/274
17q; window 274/274
>
> model17
*** Dependency models for chromosome: 17 ***
Method used: pSimCCA with window size 10
Number of models in 17p: 366, 17q: 366
Summary of dependency scores:
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.009852 0.021880 0.028740 0.039430 0.037140 0.263600
******************************************
>
> ## Information of the dependency model which has the highest dependency score
> topGenes(model17, 1)
[1] "ENSG00000141738"
>
> ## Finding a dependency model by its name
> findModel(model17, "ENSG00000129250")
*** matched case Wx = Wy with unconstrained W. Check covariances from parameters. dependency model for window size: 10 ***
Gene: ENSG00000129250 Location: 17p, 4.8686Mbp
Dependency score: 0.02380812
- WX: [1:10, 1:1] 0.0389 -0.0559 0.0238 -0.0679 ...
- WY: [1:10, 1:1] 0.0389 -0.0559 0.0238 -0.0679 ...
- phiX: [1:10, 1:10] 0.04592 0.01705 -0.01154 0.00432 ...
- phiY: [1:10, 1:10] 0.1835 0.0244 0.0222 -0.0842 ...
************************************************
>
> ## Information of the first dependency model
> model17[[1]]
*** matched case Wx = Wy with unconstrained W. Check covariances from parameters. dependency model for window size: 10 ***
Gene: ENSG00000108953 Location: 17p, 1.1949Mbp
Dependency score: 0.02688904
- WX: [1:10, 1:1] -0.0330 0.0788 -0.0572 0.1068 ...
- WY: [1:10, 1:1] -0.0330 0.0788 -0.0572 0.1068 ...
- phiX: [1:10, 1:10] 0.0917 -0.0121 -0.0148 0.0471 ...
- phiY: [1:10, 1:10] 0.3057 0.0313 -0.0998 0.0445 ...
************************************************
>
> #Plotting
> plot(model17)
>
> # genes in p arm with the highest dependency scores
> topGenes(model17[['p']], 5)
[1] "ENSG00000179094,ENSG00000179036" "ENSG00000179036"
[3] "ENSG00000178921" "ENSG00000170175"
[5] "ENSG00000133028"
>
>
>
>
>
>
> dev.off()
null device
1
>
|