Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Class "GeneDependencyModel"DescriptionA Genomic Dependency model for two data sets Objects from the ClassUsed to represent individual dependency models for screening inside ChromosomeModels. Slots
ExtendsClass DependencyModel directly. Methods
Author(s)Olli-Pekka Huovilainen ohuovila@gmail.com See AlsoFor calculation of dependency models for chromosomal arm, chromosome or
genome: Examplesdata(chromosome17) # First genomic dependency model from screening chromosomal arm models <- screen.cgh.mrna(geneExp, geneCopyNum, 10, chr=17, arm='p') model <- models[[1]] # Printing information of the model model # Latent variable Z getZ(model, geneExp,geneCopyNum) # Contributions of samples and variables to model plot(model,geneExp,geneCopyNum) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(pint)
Loading required package: mvtnorm
Loading required package: Matrix
Loading required package: dmt
Loading required package: MASS
dmt Copyright (C) 2008-2013 Leo Lahti and Olli-Pekka Huovilainen.
This program comes with ABSOLUTELY NO
WARRANTY.
This is free software, and you are welcome to redistribute it
under the FreeBSD license.
pint Copyright (C) 2008-2013 Olli-Pekka Huovilainen and Leo Lahti.
This program comes with ABSOLUTELY NO WARRANTY.
This is free software, and you are welcome to redistribute it
under the FreeBSD license.
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/pint/GeneDependencyModel-class.Rd_%03d_medium.png", width=480, height=480)
> ### Name: GeneDependencyModel-class
> ### Title: Class "GeneDependencyModel"
> ### Aliases: GeneDependencyModel-class setLoc<- setGeneName<-
> ### setChromosome<- setArm<- getScore getLoc
> ### setLoc<-,GeneDependencyModel-method
> ### setGeneName<-,GeneDependencyModel-method
> ### setChromosome<-,GeneDependencyModel-method
> ### setArm<-,GeneDependencyModel-method
> ### getScore,GeneDependencyModel-method getLoc,GeneDependencyModel-method
> ### getGeneName,GeneDependencyModel-method
> ### getWindowSize,GeneDependencyModel-method
> ### getChromosome,GeneDependencyModel-method
> ### getArm,GeneDependencyModel-method getZ,GeneDependencyModel-method
> ### Keywords: classes
>
> ### ** Examples
>
> data(chromosome17)
>
> # First genomic dependency model from screening chromosomal arm
> models <- screen.cgh.mrna(geneExp, geneCopyNum, 10, chr=17, arm='p')
Imputing missing values..
Imputing missing values..
Matching probes between the data sets..
Calculating dependency models for 17p with method pSimCCA, window size:10
17p; window 1/92
17p; window 2/92
17p; window 3/92
17p; window 4/92
17p; window 5/92
17p; window 6/92
17p; window 7/92
17p; window 8/92
17p; window 9/92
17p; window 10/92
17p; window 11/92
17p; window 12/92
17p; window 13/92
17p; window 14/92
17p; window 15/92
17p; window 16/92
17p; window 17/92
17p; window 18/92
17p; window 19/92
17p; window 20/92
17p; window 21/92
17p; window 22/92
17p; window 23/92
17p; window 24/92
17p; window 25/92
17p; window 26/92
17p; window 27/92
17p; window 28/92
17p; window 29/92
17p; window 30/92
17p; window 31/92
17p; window 32/92
17p; window 33/92
17p; window 34/92
17p; window 35/92
17p; window 36/92
17p; window 37/92
17p; window 38/92
17p; window 39/92
17p; window 40/92
17p; window 41/92
17p; window 42/92
17p; window 43/92
17p; window 44/92
17p; window 45/92
17p; window 46/92
17p; window 47/92
17p; window 48/92
17p; window 49/92
17p; window 50/92
17p; window 51/92
17p; window 52/92
17p; window 53/92
17p; window 54/92
17p; window 55/92
17p; window 56/92
17p; window 57/92
17p; window 58/92
17p; window 59/92
17p; window 60/92
17p; window 61/92
17p; window 62/92
17p; window 63/92
17p; window 64/92
17p; window 65/92
17p; window 66/92
17p; window 67/92
17p; window 68/92
17p; window 69/92
17p; window 70/92
17p; window 71/92
17p; window 72/92
17p; window 73/92
17p; window 74/92
17p; window 75/92
17p; window 76/92
17p; window 77/92
17p; window 78/92
17p; window 79/92
17p; window 80/92
17p; window 81/92
17p; window 82/92
17p; window 83/92
17p; window 84/92
17p; window 85/92
17p; window 86/92
17p; window 87/92
17p; window 88/92
17p; window 89/92
17p; window 90/92
17p; window 91/92
17p; window 92/92
> model <- models[[1]]
>
> # Printing information of the model
> model
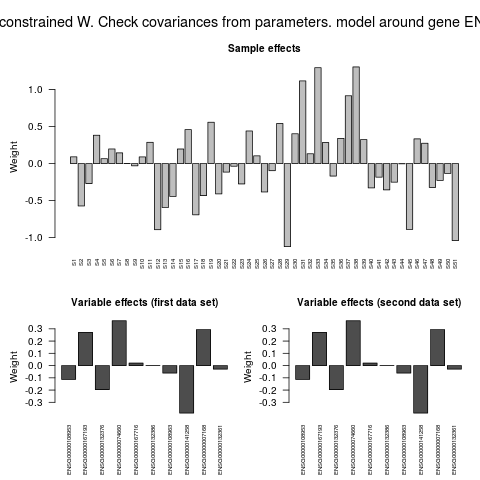
*** matched case Wx = Wy with unconstrained W. Check covariances from parameters. dependency model for window size: 10 ***
Gene: ENSG00000108953 Location: 17p, 1.1949Mbp
Dependency score: 0.02688904
- WX: [1:10, 1:1] -0.0330 0.0788 -0.0572 0.1068 ...
- WY: [1:10, 1:1] -0.0330 0.0788 -0.0572 0.1068 ...
- phiX: [1:10, 1:10] 0.0917 -0.0121 -0.0148 0.0471 ...
- phiY: [1:10, 1:10] 0.3057 0.0313 -0.0998 0.0445 ...
************************************************
>
> # Latent variable Z
> getZ(model, geneExp,geneCopyNum)
S1 S2 S3 S4 S5 S6 S7
[1,] 0.4516152 -0.669576 -0.1869619 0.2215708 -0.1391644 -0.6045729 0.2544856
S8 S9 S10 S11 S12 S13 S14
[1,] 0.2913774 -0.3416143 0.02128057 0.6240733 -1.562507 0.2730049 -0.6781598
S15 S16 S17 S18 S19 S20 S21
[1,] 0.09662121 0.2918506 -0.2893092 -0.03086981 0.3331657 0.4022143 -0.3155861
S22 S23 S24 S25 S26 S27
[1,] 0.057567 -0.02290406 0.06943918 0.02649978 -0.3570312 -0.06747664
S28 S29 S30 S31 S32 S33 S34
[1,] 0.6738301 -1.016544 0.2332853 1.239983 0.2390863 1.00107 1.197374
S35 S36 S37 S38 S39 S40 S41
[1,] -0.2065238 -0.4542886 0.5166617 0.2484803 1.448722 -0.3500769 -0.03297817
S42 S43 S44 S45 S46 S47
[1,] -0.2194966 0.06724493 0.06934309 -0.6142182 -0.04072268 0.1662846
S48 S49 S50 S51
[1,] -0.533165 -0.540564 0.08250099 -1.32432
>
> # Contributions of samples and variables to model
> plot(model,geneExp,geneCopyNum)
>
>
>
>
>
> dev.off()
null device
1
>
|