Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Class "CCProfile"DescriptionS4 class for representing coiled coil prediction results Objects from the ClassIn principle, objects of this class can be created by calls of
the form SlotsThis class extends the class
Prediction profilesAs described in f(x)=b+sum over all si(x) for i=1,… L, where L is the length of the sequence x. The values
si(x) can then be understood as the contributions that
the i-th residue makes to the overall classification of the sequence
x, which we call prediction profile. These profiles can
either be visualized as they are without taking the offset b
into account or by distributing b equally over all residues.
These are the so-called baselines that are included in
Methods
Accessor-like methodsThe
Author(s)Ulrich Bodenhofer bodenhofer@bioinf.jku.at Referenceshttp://www.bioinf.jku.at/software/procoil/ Mahrenholz, C.C., Abfalter, I.G., Bodenhofer, U., Volkmer, R., and Hochreiter, S. (2011) Complex networks govern coiled coil oligomerization - predicting and profiling by means of a machine learning approach. Mol. Cell. Proteomics 10(5):M110.004994. DOI: 10.1074/mcp.M110.004994 Palme, J., Hochreiter, S., and Bodenhofer, U. (2015) KeBABS: an R package for kernel-based analysis of biological sequences. Bioinformatics 31(15):2574-2576. DOI: 10.1093/bioinformatics/btv176 See Also
Examples
showClass("CCProfile")
## predict oligomerization of GCN4 wildtype
GCN4wt <- predict(PrOCoilModel,
"MKQLEDKVEELLSKNYHLENEVARLKKLV",
"abcdefgabcdefgabcdefgabcdefga")
## display summary of result
GCN4wt
## show raw prediction profile
profile(GCN4wt)
## plot profile
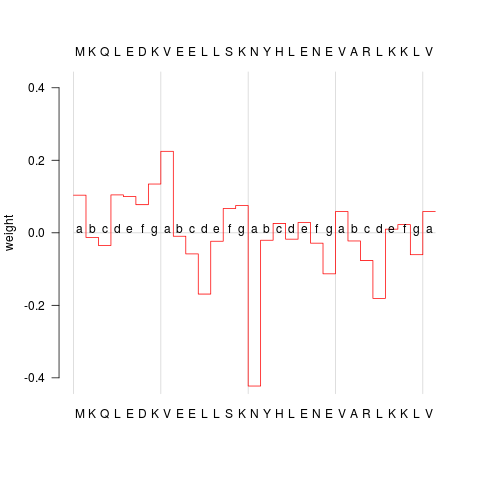
plot(GCN4wt)
## define four GCN4 mutations
GCN4mSeq <- c("GCN4wt" ="MKQLEDKVEELLSKNYHLENEVARLKKLV",
"GCN4_N16Y_L19T"="MKQLEDKVEELLSKYYHTENEVARLKKLV",
"GCN4_E22R_K27E"="MKQLEDKVEELLSKNYHLENRVARLEKLV",
"GCN4_V23K_K27E"="MKQLEDKVEELLSKNYHLENEKARLEKLV")
GCN4mReg <- rep("abcdefgabcdefgabcdefgabcdefga", 4)
## predict oligomerization
GCN4mut <- predict(PrOCoilModel, GCN4mSeq, GCN4mReg)
## display summary of result
GCN4mut
## display predictions
fitted(GCN4mut)
## overlay plot of two profiles
plot(GCN4mut[c(1, 2)])
## show heatmap
heatmap(GCN4mut)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(procoil)
Loading required package: kebabs
Loading required package: Biostrings
Loading required package: BiocGenerics
Loading required package: parallel
Attaching package: 'BiocGenerics'
The following objects are masked from 'package:parallel':
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLB
The following objects are masked from 'package:stats':
IQR, mad, xtabs
The following objects are masked from 'package:base':
Filter, Find, Map, Position, Reduce, anyDuplicated, append,
as.data.frame, cbind, colnames, do.call, duplicated, eval, evalq,
get, grep, grepl, intersect, is.unsorted, lapply, lengths, mapply,
match, mget, order, paste, pmax, pmax.int, pmin, pmin.int, rank,
rbind, rownames, sapply, setdiff, sort, table, tapply, union,
unique, unsplit
Loading required package: S4Vectors
Loading required package: stats4
Attaching package: 'S4Vectors'
The following objects are masked from 'package:base':
colMeans, colSums, expand.grid, rowMeans, rowSums
Loading required package: IRanges
Loading required package: XVector
Loading required package: kernlab
Attaching package: 'kernlab'
The following object is masked from 'package:Biostrings':
type
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/procoil/CCProfile-class.Rd_%03d_medium.png", width=480, height=480)
> ### Name: CCProfile-class
> ### Title: Class "CCProfile"
> ### Aliases: CCProfile-class CCProfile [,CCProfile,index,ANY,ANY-method
> ### show,CCProfile-method heatmap,CCProfile,missing-method
> ### baselines,CCProfile-method profiles,CCProfile-method
> ### profile,CCProfile-method sequences,CCProfile-method
> ### fitted,CCProfile-method show.CCProfile heatmap.CCProfile
> ### baselines.CCProfile profiles.CCProfile profile.CCProfile
> ### sequences.CCProfile fitted.CCProfile
> ### Keywords: classes
>
> ### ** Examples
>
> showClass("CCProfile")
Class "CCProfile" [package "procoil"]
Slots:
Name: disc pred sequences baselines profiles kernel
Class: numeric factor ANY numeric ANY ANY
Extends: "PredictionProfile"
>
> ## predict oligomerization of GCN4 wildtype
> GCN4wt <- predict(PrOCoilModel,
+ "MKQLEDKVEELLSKNYHLENEVARLKKLV",
+ "abcdefgabcdefgabcdefgabcdefga")
>
> ## display summary of result
> GCN4wt
An object of class "CCProfile"
Sequence:
A AAVector instance of length 1
width seq
[1] 29 MKQLEDKVEELLSKNYHLENEVARLKKLV
gappy pair kernel: k=1, m=5, annSpec=TRUE
Baseline: 0.03698699
Profile:
Pos 1 Pos 2 Pos 28 Pos 29
[1] 0.140762197 0.024184153 ... -0.023390414 0.095688066
Predictions:
Score Class
[1] -0.158713692 dimer
>
> ## show raw prediction profile
> profile(GCN4wt)
Pos 1 Pos 2 Pos 3 Pos 4 Pos 5 Pos 6 Pos 7
[1,] 0.1407622 0.02418415 0.002011822 0.1414524 0.1369286 0.1145901 0.1714275
Pos 8 Pos 9 Pos 10 Pos 11 Pos 12 Pos 13 Pos 14
[1,] 0.2617676 0.02745403 -0.0211955 -0.1319729 0.01365944 0.1039332 0.1122011
Pos 15 Pos 16 Pos 17 Pos 18 Pos 19 Pos 20
[1,] -0.3855509 0.01650569 0.06290137 0.01949051 0.06539256 0.00837083
Pos 21 Pos 22 Pos 23 Pos 24 Pos 25 Pos 26
[1,] -0.07611438 0.09560804 0.01451723 -0.03949032 -0.1438368 0.04703749
Pos 27 Pos 28 Pos 29
[1,] 0.05957645 -0.02339041 0.09568807
>
> ## plot profile
> plot(GCN4wt)
>
> ## define four GCN4 mutations
> GCN4mSeq <- c("GCN4wt" ="MKQLEDKVEELLSKNYHLENEVARLKKLV",
+ "GCN4_N16Y_L19T"="MKQLEDKVEELLSKYYHTENEVARLKKLV",
+ "GCN4_E22R_K27E"="MKQLEDKVEELLSKNYHLENRVARLEKLV",
+ "GCN4_V23K_K27E"="MKQLEDKVEELLSKNYHLENEKARLEKLV")
> GCN4mReg <- rep("abcdefgabcdefgabcdefgabcdefga", 4)
>
> ## predict oligomerization
> GCN4mut <- predict(PrOCoilModel, GCN4mSeq, GCN4mReg)
>
> ## display summary of result
> GCN4mut
An object of class "CCProfile"
Sequences:
A AAVector instance of length 4
width seq names
[1] 29 MKQLEDKVEELLSKNYHLENEVARLKKLV GCN4wt
[2] 29 MKQLEDKVEELLSKYYHTENEVARLKKLV GCN4_N16Y_L19T
[3] 29 MKQLEDKVEELLSKNYHLENRVARLEKLV GCN4_E22R_K27E
[4] 29 MKQLEDKVEELLSKNYHLENEKARLEKLV GCN4_V23K_K27E
gappy pair kernel: k=1, m=5, annSpec=TRUE
Baselines: 0.03698699 0.03698699 0.03698699 0.03698699
Profiles:
Pos 1 Pos 2 Pos 28 Pos 29
GCN4wt 0.140762197 0.024184153 ... -0.023390414 0.095688066
GCN4_N16Y_L19T 0.144175109 0.024770521 ... -0.023957537 0.098008113
GCN4_E22R_K27E 0.137580719 0.023637548 ... -0.042715408 0.148626188
GCN4_V23K_K27E 0.141592659 0.024326834 ... -0.047527521 0.152960221
Predictions:
Score Class
GCN4wt -0.158713692 dimer
GCN4_N16Y_L19T 0.420763995 trimer
GCN4_E22R_K27E 0.623458294 trimer
GCN4_V23K_K27E -0.500406810 dimer
>
> ## display predictions
> fitted(GCN4mut)
GCN4wt GCN4_N16Y_L19T GCN4_E22R_K27E GCN4_V23K_K27E
dimer trimer trimer dimer
Levels: dimer trimer
>
> ## overlay plot of two profiles
> plot(GCN4mut[c(1, 2)])
>
> ## show heatmap
> heatmap(GCN4mut)
>
>
>
>
>
> dev.off()
null device
1
>
|