Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Fit a semiparametric regression model with spatially adaptive penalized splinesDescriptionFits semiparametric
regression models using the mixed model
representation of penalized splines with spatially adaptive
penalties based on the Usage
asp2(form, spar.method = "REML", contrasts=NULL,
omit.missing = NULL, returnFit=FALSE,
niter = 20, niter.var = 50, tol=1e-6, tol.theta=1e-6,
control=NULL)
Arguments
ValueA list object of class
Author(s)Manuel Wiesenfarth m.wiesenfarth at dkfz de, Tatyana Krivobokova tkrivob at gwdg.de ReferencesKrivobokova, T., Crainiceanu, C.M. and Kauermann, G. (2008) Ruppert, D., Wand, M.P. and Carroll, R.J. (2003) Wiesenfarth, M., Krivobokova, T., Klasen, S., Sperlich, S. (2012). See Also
Examples
############
# Examples as in package AdaptFit
## scatterplot smoothing
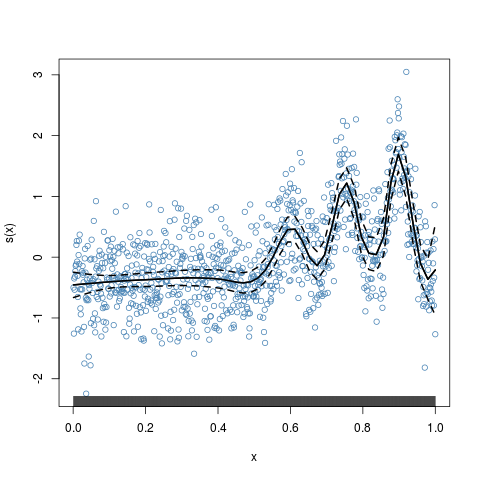
x <- 1:1000/1000
mu <- exp(-400*(x-0.6)^2)+
5*exp(-500*(x-0.75)^2)/3+2*exp(-500*(x-0.9)^2)
y <- mu+0.5*rnorm(1000)
#fit with default knots
y.fit <- asp2(y~f(x,adap=TRUE))
plot(y.fit,residuals=TRUE,lwd=2,scb.lwd=2,scb.lty="dashed")
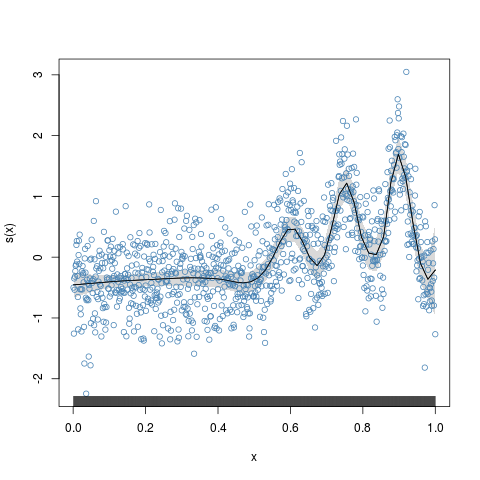
# with shaded confidence region.
# Use scb.lty="blank" to plot the shades only.
plot(y.fit,residuals=TRUE,shade=TRUE,scb.lty="blank")
## Not run:
## additive models
x1 <- 1:300/300
x2 <- runif(300)
mu1 <- exp(-400*(x1-0.6)^2)+
5*exp(-500*(x1-0.75)^2)/3+2*exp(-500*(x1-0.9)^2)
mu2 <- sin(2*pi*x2)
y2 <- mu1+mu2+0.3*rnorm(300)
y2.fit <- asp2(y2~f(x1,adap=TRUE)+f(x2,adap=TRUE))
# switch off adaptive fitting for the first function
y21.fit <- asp2(y2~f(x1,adap=FALSE)+f(x2,adap=TRUE))
op <- par(mfrow = c(2, 2))
plot(y2.fit)
plot(y21.fit)
par(op)
## scatterplot smoothing with specified knots and subknots
x <- 1:400/400
mu <- sqrt(x*(1-x))*sin((2*pi*(1+2^((9-4*6)/5)))/(x+2^((9-4*6)/5)))
y <- mu+0.2*rnorm(400)
kn <- default.knots(x,80)
kn.var <- default.knots(kn,20)
y.fit <- asp2(y~f(x,knots=kn))
y.fit2 <- asp2(y~f(x,knots=kn,var.knots=kn.var,adap=TRUE))
op <- par(mfrow = c(1, 2))
plot(y.fit)
plot(y.fit2)
par(op)
##################
#more examples
beta=function(l,m,x)
return(gamma(l+m)*(gamma(l)*gamma(m))^(-1)*x^(l-1)*(1-x)^(m-1))
f1 = function(x) return((0.6*beta(30,17,x)+0.4*beta(3,11,x))*1/0.958)
f2 = function(z) return((sin(2*pi*(z-0.5))^2)*1/.3535)
f3 = function(z)
return((exp(-400*(z-0.6)^2)+
5/3*exp(-500*(z-0.75)^2)+2*exp(-500*(z-0.9)^2))*1/0.549)
set.seed(1)
N <- 500
x1 = runif(N,0,1)
x2 = runif(N,0,1)
x3 = runif(N,0,1)
kn1 <- default.knots(x1,40)
kn2 <- default.knots(x2,40)
kn3 <- default.knots(x3,40)
kn.var3 <- default.knots(kn3,5)
y <- f1(x1)+f2(x2)+f3(x3)+0.3*rnorm(N)
# semiparametric model
fit1= asp2(y~x1+f(x2,basis="os",degree=3,knots=kn2,adap=FALSE)
+f(x3,basis="os",degree=3,
knots=kn3,var.knots=kn.var3,adap=FALSE),
niter = 20, niter.var = 200)
summary(fit1)
plot(fit1,pages=1)
# all effects flexible
# fit model with all smoothing parameters constant
fit2a= asp2(y~f(x1,basis="os",degree=3,knots=kn1,adap=FALSE)
+f(x2,basis="os",degree=3,knots=kn2,adap=FALSE)
+f(x3,basis="os",degree=3,knots=kn3,adap=FALSE),
niter = 20, niter.var = 200)
plot(fit2a,pages=1)
# fit model with last smoothing parameter adaptive
fit2b= asp2(y~f(x1,basis="os",degree=3,knots=kn1,adap=FALSE)
+f(x2,basis="os",degree=3,knots=kn2,adap=FALSE)
+f(x3,basis="os",degree=3,knots=kn3,adap=TRUE,
var.knots=kn.var3,var.basis="os",var.degree=3),
niter = 20, niter.var = 200)
# plot smoothing parameter function for covariate x3.
# Note that in the case of B-splines additional knots are added,
# see references.
plot(seq(0,1,length.out=42), fit2b$y.cov/fit2b$random.var[85:126],
ylab=expression(lambda(x3)),xlab="x3",type="l",lwd=3)
# compute 95
# You could skip this and use "fit2b" indstead of "scb2b" later on, however,
# if N is large, computing the SCBs various times can take some time
# if you don't need fitted values and bounds for all covariate points
# (can be computationally intensive due to large matrix dimensions),
# set calc.stdev=F such that these are not computed.
scb2b<- scbM(fit2b,calc.stdev=FALSE)
plot(scb2b,pages=1)
# plot only f(x2).
plot(scb2b,select=2,mfrow=c(1,1),lwd=3,ylab="f(x2)",xlab="x2")
# plot.scbm (and plot.asp) returns fitted values and confidence limits,
# if you only need the returned object set plot=FALSE
pscb=plot(scb2b,plot=FALSE)
# add pointwise confidence intervals to the plot
polygon(c(pscb$grid.x[[2]], rev(pscb$grid.x[[2]])),
c(pscb$fitted[[2]]+1.96*pscb$Stdev[[2]],
rev(pscb$fitted[[2]]-1.96*pscb$Stdev[[2]])),
col = grey(0.85), border = NA)
lines(pscb$grid.x[[2]],pscb$lcb[[2]],lty="dotted",lwd=3)
lines(pscb$grid.x[[2]],pscb$fitted[[2]],lwd=3)
lines(pscb$grid.x[[2]],pscb$ucb[[2]],lty="dotted",lwd=3)
# plot first derivative of f(x1)
scb2bdrv<- scbM(fit2b,drv=1,calc.stdev=FALSE)
plot(scb2bdrv,select=1)
#the following would give the same result
#x11();plot(fit2b,select=1,drv=1)
# different style
plot(scb2bdrv,select=1,scb.lty="blank",
shade=TRUE,shade.col="steelblue")
## End(Not run)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(AdaptFitOS)
Loading required package: nlme
Loading required package: MASS
Loading required package: splines
AdaptFitOS 0.62 loaded. Type 'help("AdaptFitOS-package")' for an overview.
Please cite as:
Wiesenfarth, M., Krivobokova, T., Klasen, S., & Sperlich, S. (2012).
Direct simultaneous inference in additive models and its application to model undernutrition.
Journal of the American Statistical Association, 107(500), 1286-1296.
Attaching package: 'AdaptFitOS'
The following object is masked from 'package:stats':
sigma
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/AdaptFitOS/asp2.Rd_%03d_medium.png", width=480, height=480)
> ### Name: asp2
> ### Title: Fit a semiparametric regression model with spatially adaptive
> ### penalized splines
> ### Aliases: asp2
> ### Keywords: nonlinear models smooth regression adaptive
>
> ### ** Examples
>
> ############
> # Examples as in package AdaptFit
> ## scatterplot smoothing
> x <- 1:1000/1000
> mu <- exp(-400*(x-0.6)^2)+
+ 5*exp(-500*(x-0.75)^2)/3+2*exp(-500*(x-0.9)^2)
> y <- mu+0.5*rnorm(1000)
>
> #fit with default knots
> y.fit <- asp2(y~f(x,adap=TRUE))
> plot(y.fit,residuals=TRUE,lwd=2,scb.lwd=2,scb.lty="dashed")
Critical value for f(x): 3.423244
> # with shaded confidence region.
> # Use scb.lty="blank" to plot the shades only.
> plot(y.fit,residuals=TRUE,shade=TRUE,scb.lty="blank")
Critical value for f(x): 3.423244
>
> ## Not run:
> ##D ## additive models
> ##D x1 <- 1:300/300
> ##D x2 <- runif(300)
> ##D mu1 <- exp(-400*(x1-0.6)^2)+
> ##D 5*exp(-500*(x1-0.75)^2)/3+2*exp(-500*(x1-0.9)^2)
> ##D mu2 <- sin(2*pi*x2)
> ##D y2 <- mu1+mu2+0.3*rnorm(300)
> ##D
> ##D y2.fit <- asp2(y2~f(x1,adap=TRUE)+f(x2,adap=TRUE))
> ##D # switch off adaptive fitting for the first function
> ##D y21.fit <- asp2(y2~f(x1,adap=FALSE)+f(x2,adap=TRUE))
> ##D op <- par(mfrow = c(2, 2))
> ##D plot(y2.fit)
> ##D plot(y21.fit)
> ##D par(op)
> ##D
> ##D
> ##D ## scatterplot smoothing with specified knots and subknots
> ##D x <- 1:400/400
> ##D mu <- sqrt(x*(1-x))*sin((2*pi*(1+2^((9-4*6)/5)))/(x+2^((9-4*6)/5)))
> ##D y <- mu+0.2*rnorm(400)
> ##D
> ##D kn <- default.knots(x,80)
> ##D kn.var <- default.knots(kn,20)
> ##D
> ##D y.fit <- asp2(y~f(x,knots=kn))
> ##D y.fit2 <- asp2(y~f(x,knots=kn,var.knots=kn.var,adap=TRUE))
> ##D op <- par(mfrow = c(1, 2))
> ##D plot(y.fit)
> ##D plot(y.fit2)
> ##D par(op)
> ##D
> ##D ##################
> ##D #more examples
> ##D beta=function(l,m,x)
> ##D return(gamma(l+m)*(gamma(l)*gamma(m))^(-1)*x^(l-1)*(1-x)^(m-1))
> ##D f1 = function(x) return((0.6*beta(30,17,x)+0.4*beta(3,11,x))*1/0.958)
> ##D f2 = function(z) return((sin(2*pi*(z-0.5))^2)*1/.3535)
> ##D f3 = function(z)
> ##D return((exp(-400*(z-0.6)^2)+
> ##D 5/3*exp(-500*(z-0.75)^2)+2*exp(-500*(z-0.9)^2))*1/0.549)
> ##D
> ##D set.seed(1)
> ##D N <- 500
> ##D x1 = runif(N,0,1)
> ##D x2 = runif(N,0,1)
> ##D x3 = runif(N,0,1)
> ##D
> ##D
> ##D kn1 <- default.knots(x1,40)
> ##D kn2 <- default.knots(x2,40)
> ##D kn3 <- default.knots(x3,40)
> ##D kn.var3 <- default.knots(kn3,5)
> ##D
> ##D y <- f1(x1)+f2(x2)+f3(x3)+0.3*rnorm(N)
> ##D
> ##D # semiparametric model
> ##D fit1= asp2(y~x1+f(x2,basis="os",degree=3,knots=kn2,adap=FALSE)
> ##D +f(x3,basis="os",degree=3,
> ##D knots=kn3,var.knots=kn.var3,adap=FALSE),
> ##D niter = 20, niter.var = 200)
> ##D summary(fit1)
> ##D plot(fit1,pages=1)
> ##D
> ##D
> ##D # all effects flexible
> ##D # fit model with all smoothing parameters constant
> ##D fit2a= asp2(y~f(x1,basis="os",degree=3,knots=kn1,adap=FALSE)
> ##D +f(x2,basis="os",degree=3,knots=kn2,adap=FALSE)
> ##D +f(x3,basis="os",degree=3,knots=kn3,adap=FALSE),
> ##D niter = 20, niter.var = 200)
> ##D plot(fit2a,pages=1)
> ##D
> ##D # fit model with last smoothing parameter adaptive
> ##D fit2b= asp2(y~f(x1,basis="os",degree=3,knots=kn1,adap=FALSE)
> ##D +f(x2,basis="os",degree=3,knots=kn2,adap=FALSE)
> ##D +f(x3,basis="os",degree=3,knots=kn3,adap=TRUE,
> ##D var.knots=kn.var3,var.basis="os",var.degree=3),
> ##D niter = 20, niter.var = 200)
> ##D
> ##D # plot smoothing parameter function for covariate x3.
> ##D # Note that in the case of B-splines additional knots are added,
> ##D # see references.
> ##D plot(seq(0,1,length.out=42), fit2b$y.cov/fit2b$random.var[85:126],
> ##D ylab=expression(lambda(x3)),xlab="x3",type="l",lwd=3)
> ##D
> ##D # compute 95##D
> ##D # You could skip this and use "fit2b" indstead of "scb2b" later on, however,
> ##D # if N is large, computing the SCBs various times can take some time
> ##D # if you don't need fitted values and bounds for all covariate points
> ##D # (can be computationally intensive due to large matrix dimensions),
> ##D # set calc.stdev=F such that these are not computed.
> ##D scb2b<- scbM(fit2b,calc.stdev=FALSE)
> ##D plot(scb2b,pages=1)
> ##D
> ##D # plot only f(x2).
> ##D plot(scb2b,select=2,mfrow=c(1,1),lwd=3,ylab="f(x2)",xlab="x2")
> ##D # plot.scbm (and plot.asp) returns fitted values and confidence limits,
> ##D # if you only need the returned object set plot=FALSE
> ##D pscb=plot(scb2b,plot=FALSE)
> ##D # add pointwise confidence intervals to the plot
> ##D polygon(c(pscb$grid.x[[2]], rev(pscb$grid.x[[2]])),
> ##D c(pscb$fitted[[2]]+1.96*pscb$Stdev[[2]],
> ##D rev(pscb$fitted[[2]]-1.96*pscb$Stdev[[2]])),
> ##D col = grey(0.85), border = NA)
> ##D lines(pscb$grid.x[[2]],pscb$lcb[[2]],lty="dotted",lwd=3)
> ##D lines(pscb$grid.x[[2]],pscb$fitted[[2]],lwd=3)
> ##D lines(pscb$grid.x[[2]],pscb$ucb[[2]],lty="dotted",lwd=3)
> ##D
> ##D # plot first derivative of f(x1)
> ##D scb2bdrv<- scbM(fit2b,drv=1,calc.stdev=FALSE)
> ##D plot(scb2bdrv,select=1)
> ##D #the following would give the same result
> ##D #x11();plot(fit2b,select=1,drv=1)
> ##D # different style
> ##D plot(scb2bdrv,select=1,scb.lty="blank",
> ##D shade=TRUE,shade.col="steelblue")
> ## End(Not run)
>
>
>
>
>
> dev.off()
null device
1
>
|