Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Bayesian Estimation Supersedes the t TestDescriptionAn alternative to t tests, producing posterior estimates for groups means and standard deviations and their differences and effect sizes. Bayesian estimation provides a much richer picture of the data, and can be summarised as point estimates and credible intervals. DetailsThe core function, In addition, the procedure accounts for departures from normality by using a t-distribution to model the variable of interest and estimating a measure of normality. Functions to summarise and to visualise the output are provided. The function Author(s)Original code by John K. Kruschke, http://www.indiana.edu/~kruschke/BEST/ Maintainer: Mike Meredith <mmeredith at wcs.org> ReferencesKruschke, J. K. 2013. Bayesian estimation supersedes the t test. Journal of Experimental Psychology: General 142(2):573-603. doi: 10.1037/a0029146 Kruschke, J. K. 2011. Doing Bayesian data analysis: a tutorial with R and BUGS. Elsevier, Amsterdam, especially Chapter 18. Examples
## Comparison of two groups:
## =========================
y1 <- c(5.77, 5.33, 4.59, 4.33, 3.66, 4.48)
y2 <- c(3.88, 3.55, 3.29, 2.59, 2.33, 3.59)
# Run an analysis, takes up to 1 min.
BESTout <- BESTmcmc(y1, y2)
# Look at the result:
BESTout
summary(BESTout)
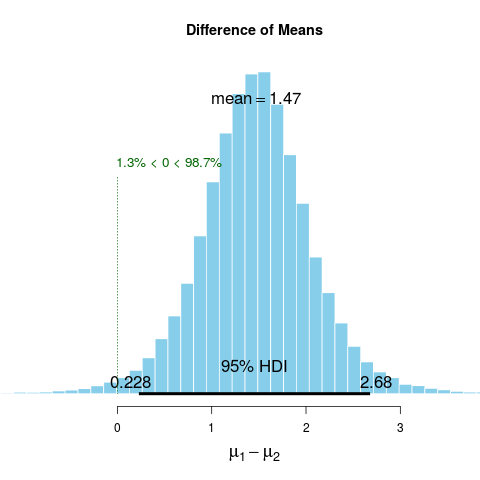
plot(BESTout)
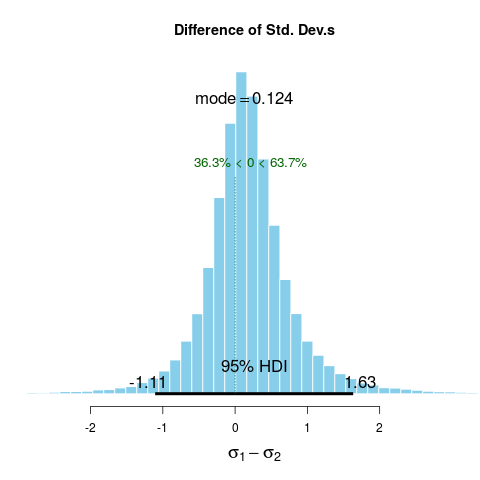
plot(BESTout, "sd")
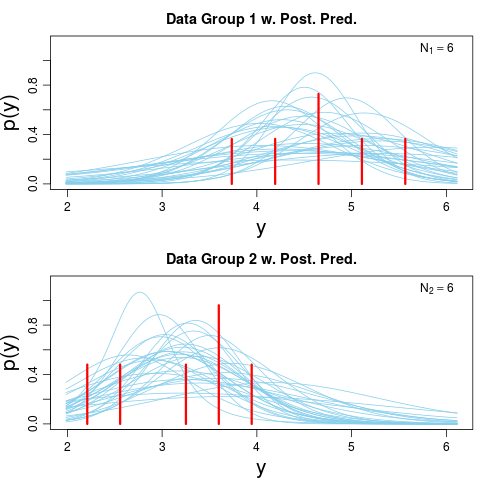
plotPostPred(BESTout)
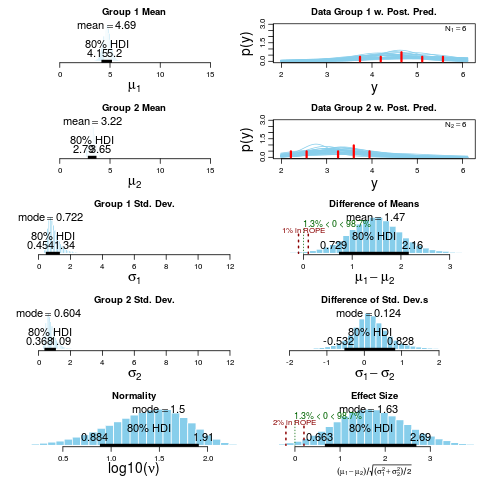
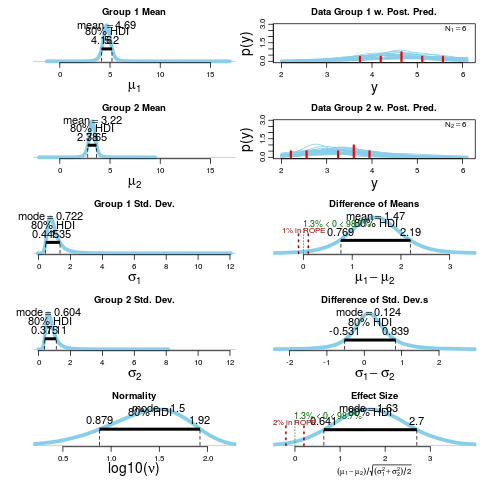
plotAll(BESTout, credMass=0.8, ROPEm=c(-0.1,0.1),
ROPEeff=c(-0.2,0.2), compValm=0.5)
plotAll(BESTout, credMass=0.8, ROPEm=c(-0.1,0.1),
ROPEeff=c(-0.2,0.2), compValm=0.5, showCurve=TRUE)
summary(BESTout, credMass=0.8, ROPEm=c(-0.1,0.1), ROPEsd=c(-0.15,0.15),
ROPEeff=c(-0.2,0.2))
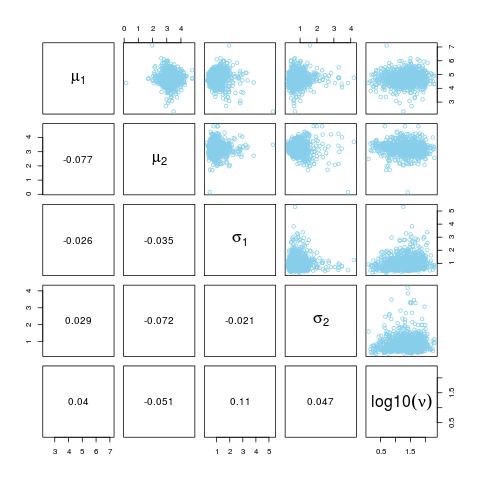
pairs(BESTout)
head(BESTout$mu1)
muDiff <- BESTout$mu1 - BESTout$mu2
mean(muDiff > 1.5)
mean(BESTout$sigma1 - BESTout$sigma2)
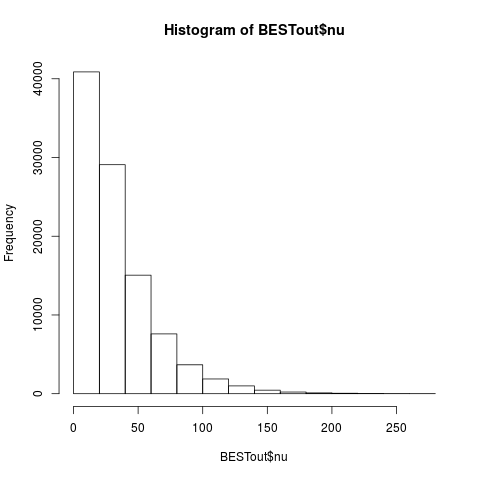
hist(BESTout$nu)
# Retrospective power analysis
# ----------------------------
# This takes time, so we do 2 simulations here; a real analysis needs several hundred
powerRet <- BESTpower(BESTout, N1=length(y1), N2=length(y2),
ROPEm=c(-0.1,0.1), maxHDIWm=2.0, nRep=2)
powerRet
# We only set criteria for the mean, so results for sd and effect size are all NA.
## Analysis with a single group:
## =============================
y0 <- c(1.89, 1.78, 1.30, 1.74, 1.33, 0.89)
# Run an analysis, takes up to 40 secs.
BESTout1 <- BESTmcmc(y0)
BESTout1
summary(BESTout1)
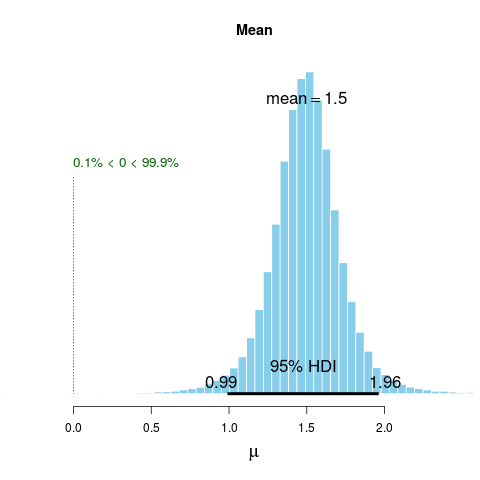
plot(BESTout1)
head(BESTout1$mu)
mean(BESTout1$sigma)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(BEST)
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/BEST/BEST-package.Rd_%03d_medium.png", width=480, height=480)
> ### Name: BEST-package
> ### Title: Bayesian Estimation Supersedes the t Test
> ### Aliases: BEST-package BEST
> ### Keywords: package htest
>
> ### ** Examples
>
>
> ## No test:
> ## Comparison of two groups:
> ## =========================
> y1 <- c(5.77, 5.33, 4.59, 4.33, 3.66, 4.48)
> y2 <- c(3.88, 3.55, 3.29, 2.59, 2.33, 3.59)
>
> # Run an analysis, takes up to 1 min.
> BESTout <- BESTmcmc(y1, y2)
Processing function input.......
Done.
Beginning parallel processing using 3 cores. Console output will be suppressed.
Parallel processing completed.
MCMC took 0.088 minutes.
>
> # Look at the result:
> BESTout
MCMC fit results for BEST analysis:
100002 simulations saved.
mean sd median HDIlo HDIup Rhat n.eff
mu1 4.6880 0.4938 4.6842 3.7416 5.623 1.006 30831
mu2 3.2166 0.3958 3.2209 2.4345 3.982 1.000 30077
nu 33.4806 29.1211 25.1112 1.0144 91.328 1.000 20431
sigma1 1.0198 0.5462 0.8845 0.3578 2.014 1.005 9622
sigma2 0.8371 0.4339 0.7290 0.3059 1.645 1.001 9122
'HDIlo' and 'HDIup' are the limits of a 95% HDI credible interval.
'Rhat' is the potential scale reduction factor (at convergence, Rhat=1).
'n.eff' is a crude measure of effective sample size.
> summary(BESTout)
mean median mode HDI% HDIlo HDIup compVal %>compVal
mu1 4.688 4.684 4.662 95 3.742 5.62
mu2 3.217 3.221 3.244 95 2.435 3.98
muDiff 1.471 1.467 1.500 95 0.228 2.68 0 98.7
sigma1 1.020 0.885 0.722 95 0.358 2.01
sigma2 0.837 0.729 0.604 95 0.306 1.64
sigmaDiff 0.183 0.148 0.124 95 -1.106 1.63 0 63.7
nu 33.481 25.111 8.858 95 1.014 91.33
log10nu 1.362 1.400 1.495 95 0.549 2.09
effSz 1.702 1.680 1.626 95 0.179 3.30 0 98.7
> plot(BESTout)
> plot(BESTout, "sd")
> plotPostPred(BESTout)
> plotAll(BESTout, credMass=0.8, ROPEm=c(-0.1,0.1),
+ ROPEeff=c(-0.2,0.2), compValm=0.5)
> plotAll(BESTout, credMass=0.8, ROPEm=c(-0.1,0.1),
+ ROPEeff=c(-0.2,0.2), compValm=0.5, showCurve=TRUE)
> summary(BESTout, credMass=0.8, ROPEm=c(-0.1,0.1), ROPEsd=c(-0.15,0.15),
+ ROPEeff=c(-0.2,0.2))
mean median mode HDI% HDIlo HDIup compVal %>compVal ROPElow
mu1 4.688 4.684 4.662 80 4.146 5.195
mu2 3.217 3.221 3.244 80 2.794 3.653
muDiff 1.471 1.467 1.500 80 0.729 2.161 0 98.7 -0.10
sigma1 1.020 0.885 0.722 80 0.454 1.335
sigma2 0.837 0.729 0.604 80 0.368 1.087
sigmaDiff 0.183 0.148 0.124 80 -0.532 0.828 0 63.7 -0.15
nu 33.481 25.111 8.858 80 1.418 52.029
log10nu 1.362 1.400 1.495 80 0.884 1.910
effSz 1.702 1.680 1.626 80 0.663 2.692 0 98.7 -0.20
ROPEhigh %InROPE
mu1
mu2
muDiff 0.10 0.666
sigma1
sigma2
sigmaDiff 0.15 25.478
nu
log10nu
effSz 0.20 1.925
> pairs(BESTout)
>
> head(BESTout$mu1)
[1] 4.616803 4.916176 4.669184 4.562231 5.138517 5.147933
> muDiff <- BESTout$mu1 - BESTout$mu2
> mean(muDiff > 1.5)
[1] 0.4755205
> mean(BESTout$sigma1 - BESTout$sigma2)
[1] 0.182755
> hist(BESTout$nu)
>
> # Retrospective power analysis
> # ----------------------------
> # This takes time, so we do 2 simulations here; a real analysis needs several hundred
>
> powerRet <- BESTpower(BESTout, N1=length(y1), N2=length(y2),
+ ROPEm=c(-0.1,0.1), maxHDIWm=2.0, nRep=2)
:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
Power computation: Simulated Experiment 1 of 2 :
Processing function input.......
Done.
Beginning parallel processing using 3 cores. Console output will be suppressed.
Parallel processing completed.
MCMC took 0.035 minutes.
After 1 Simulated Experiments, Posterior Probability
of meeting each criterion is (mean and 95% CrI):
mean CrIlo CrIhi
mean: HDI > ROPE 0.333 0 0.776
mean: HDI < ROPE 0.333 0 0.776
mean: HDI in ROPE 0.333 0 0.776
mean: HDI width ok 0.333 0 0.776
:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
Power computation: Simulated Experiment 2 of 2 :
Processing function input.......
Done.
Beginning parallel processing using 3 cores. Console output will be suppressed.
Parallel processing completed.
MCMC took 0.03 minutes.
After 2 Simulated Experiments, Posterior Probability
of meeting each criterion is (mean and 95% CrI):
mean CrIlo CrIhi
mean: HDI > ROPE 0.25 0 0.632
mean: HDI < ROPE 0.25 0 0.632
mean: HDI in ROPE 0.25 0 0.632
mean: HDI width ok 0.25 0 0.632
> powerRet
mean CrIlo CrIhi
mean: HDI > ROPE 0.25 2.114738e-09 0.6315969
mean: HDI < ROPE 0.25 2.114738e-09 0.6315969
mean: HDI in ROPE 0.25 2.114738e-09 0.6315969
mean: HDI width ok 0.25 2.114738e-09 0.6315969
sd: HDI > ROPE NA NA NA
sd: HDI < ROPE NA NA NA
sd: HDI in ROPE NA NA NA
sd: HDI width ok NA NA NA
effect: HDI > ROPE NA NA NA
effect: HDI < ROPE NA NA NA
effect: HDI in ROPE NA NA NA
effect: HDI width ok NA NA NA
> # We only set criteria for the mean, so results for sd and effect size are all NA.
>
> ## Analysis with a single group:
> ## =============================
> y0 <- c(1.89, 1.78, 1.30, 1.74, 1.33, 0.89)
>
> # Run an analysis, takes up to 40 secs.
> BESTout1 <- BESTmcmc(y0)
Processing function input.......
Done.
Beginning parallel processing using 3 cores. Console output will be suppressed.
Parallel processing completed.
MCMC took 0.074 minutes.
> BESTout1
MCMC fit results for BEST analysis:
100002 simulations saved.
mean sd median HDIlo HDIup Rhat n.eff
mu 1.4970 0.2689 1.4970 0.9901 1.963 1.014 17869
nu 32.3996 29.8412 23.3732 1.0003 91.703 1.000 18480
sigma 0.5278 0.3307 0.4518 0.1851 1.033 1.036 3865
'HDIlo' and 'HDIup' are the limits of a 95% HDI credible interval.
'Rhat' is the potential scale reduction factor (at convergence, Rhat=1).
'n.eff' is a crude measure of effective sample size.
> summary(BESTout1)
mean median mode HDI% HDIlo HDIup compVal %>compVal
mu 1.497 1.497 1.492 95 0.990 1.96 0 99.9
sigma 0.528 0.452 0.371 95 0.185 1.03
nu 32.400 23.373 7.314 95 1.000 91.70
log10nu 1.329 1.369 1.450 95 0.449 2.09
effSz 3.444 3.315 3.010 95 0.831 6.24 0 99.9
> plot(BESTout1)
>
> head(BESTout1$mu)
[1] 1.712800 1.699769 1.745293 1.797035 1.418139 1.592211
> mean(BESTout1$sigma)
[1] 0.5277566
> ## End(No test)
>
>
>
>
>
> dev.off()
null device
1
>
|