Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Bayesian Model Averaging for linear regression models.DescriptionBayesian Model Averaging accounts for the model uncertainty inherent in the variable selection problem by averaging over the best models in the model class according to approximate posterior model probability. Usage
bicreg(x, y, wt = rep(1, length(y)), strict = FALSE, OR = 20,
maxCol = 31, drop.factor.levels = TRUE, nbest = 150)
Arguments
DetailsBayesian Model Averaging accounts for the model uncertainty inherent in the variable selection problem by averaging over the best models in the model class according to the approximate posterior model probabilities. Value
The function 'summary' is used to print a summary of the results. The function 'plot' is used to plot posterior distributions for the coefficients. An object of class
Author(s)Original Splus code developed by Adrian Raftery (raftery@AT@stat.washington.edu) and revised by Chris T. Volinsky. Translation to R by Ian Painter. ReferencesRaftery, Adrian E. (1995). Bayesian model selection in social research (with Discussion). Sociological Methodology 1995 (Peter V. Marsden, ed.), pp. 111-196, Cambridge, Mass.: Blackwells. See Also
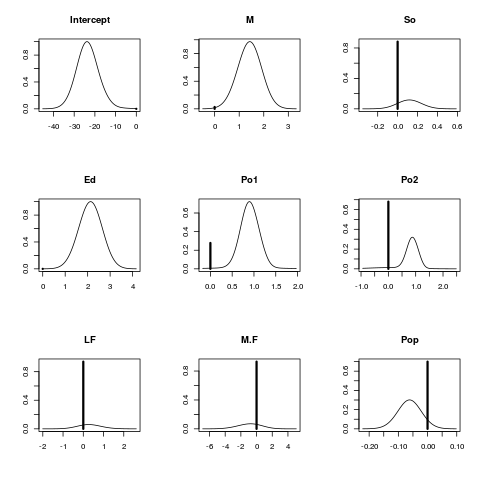
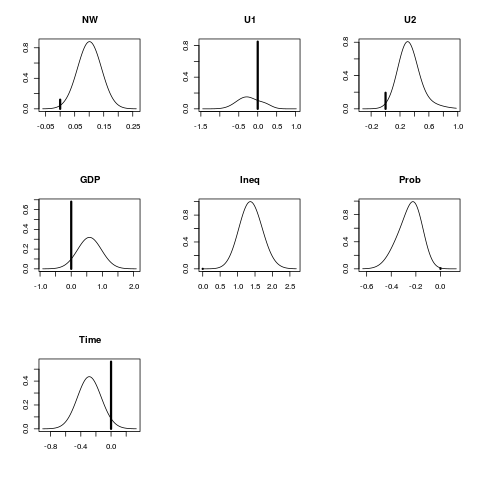
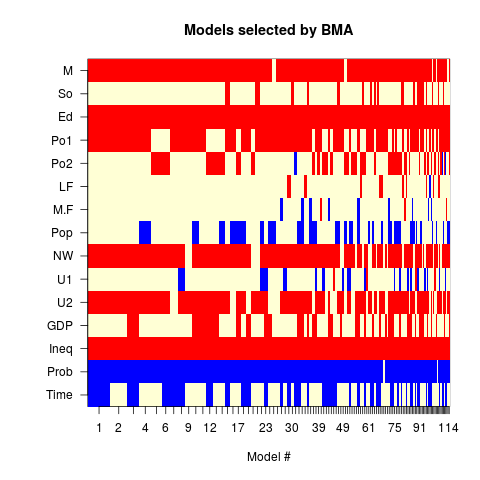
Exampleslibrary(MASS) data(UScrime) x<- UScrime[,-16] y<- log(UScrime[,16]) x[,-2]<- log(x[,-2]) lma<- bicreg(x, y, strict = FALSE, OR = 20) summary(lma) plot(lma) imageplot.bma(lma) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(BMA)
Loading required package: survival
Loading required package: leaps
Loading required package: robustbase
Attaching package: 'robustbase'
The following object is masked from 'package:survival':
heart
Loading required package: inline
Loading required package: rrcov
Scalable Robust Estimators with High Breakdown Point (version 1.3-11)
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/BMA/bicreg.Rd_%03d_medium.png", width=480, height=480)
> ### Name: bicreg
> ### Title: Bayesian Model Averaging for linear regression models.
> ### Aliases: bicreg
> ### Keywords: regression models
>
> ### ** Examples
>
> library(MASS)
> data(UScrime)
> x<- UScrime[,-16]
> y<- log(UScrime[,16])
> x[,-2]<- log(x[,-2])
> lma<- bicreg(x, y, strict = FALSE, OR = 20)
> summary(lma)
Call:
bicreg(x = x, y = y, strict = FALSE, OR = 20)
115 models were selected
Best 5 models (cumulative posterior probability = 0.2039 ):
p!=0 EV SD model 1 model 2 model 3
Intercept 100.0 -23.45301 5.58897 -22.63715 -24.38362 -25.94554
M 97.3 1.38103 0.53531 1.47803 1.51437 1.60455
So 11.7 0.01398 0.05640 . . .
Ed 100.0 2.12101 0.52527 2.22117 2.38935 1.99973
Po1 72.2 0.64849 0.46544 0.85244 0.91047 0.73577
Po2 32.0 0.24735 0.43829 . . .
LF 6.0 0.01834 0.16242 . . .
M.F 7.0 -0.06285 0.46566 . . .
Pop 30.1 -0.01862 0.03626 . . .
NW 88.0 0.08894 0.05089 0.10888 0.08456 0.11191
U1 15.1 -0.03282 0.14586 . . .
U2 80.7 0.26761 0.19882 0.28874 0.32169 0.27422
GDP 31.9 0.18726 0.34986 . . 0.54105
Ineq 100.0 1.38180 0.33460 1.23775 1.23088 1.41942
Prob 99.2 -0.24962 0.09999 -0.31040 -0.19062 -0.29989
Time 43.7 -0.12463 0.17627 -0.28659 . -0.29682
nVar 8 7 9
r2 0.842 0.826 0.851
BIC -55.91243 -55.36499 -54.69225
post prob 0.062 0.047 0.034
model 4 model 5
Intercept -22.80644 -24.50477
M 1.26830 1.46061
So . .
Ed 2.17788 2.39875
Po1 0.98597 .
Po2 . 0.90689
LF . .
M.F . .
Pop -0.05685 .
NW 0.09745 0.08534
U1 . .
U2 0.28054 0.32977
GDP . .
Ineq 1.32157 1.29370
Prob -0.21636 -0.20614
Time . .
nVar 8 7
r2 0.838 0.823
BIC -54.60434 -54.40788
post prob 0.032 0.029
> plot(lma)
>
> imageplot.bma(lma)
>
>
>
>
>
>
> dev.off()
null device
1
>
|