|
Last data update: 2014.03.03
|
R: Transform confidence intervals from glm fits.
Transform confidence intervals from glm fits.
Description
Transform confidence intervals derived from glm fits back to original scale and give appropriate names.
Usage
## S3 method for class 'glht'
UnlogCI(x)
Arguments
x |
an object of class "confint.glht"
|
Details
Applies exponential function on the estimates and confidence limits and creates useful names for the comparisons and parameters.
Value
An object of class "UnlogCI".
See Also
plotCI.UnlogCI for plotting the result
Examples
# # # CI for odds ratios
# # # for models on the logit-link
data(Feeding)
# Larval mortality:
Feeding$Lmort <- Feeding$Total - Feeding$Pupating
fit1<-glm(cbind(Pupating,Lmort)~Variety,data=Feeding, family=quasibinomial)
anova(fit1, test="F")
library(multcomp)
comp<-glht(fit1, mcp(Variety="Tukey"))
CIraw<-CIGLM(comp,method="Raw")
CIraw
UnlogCI(CIraw)
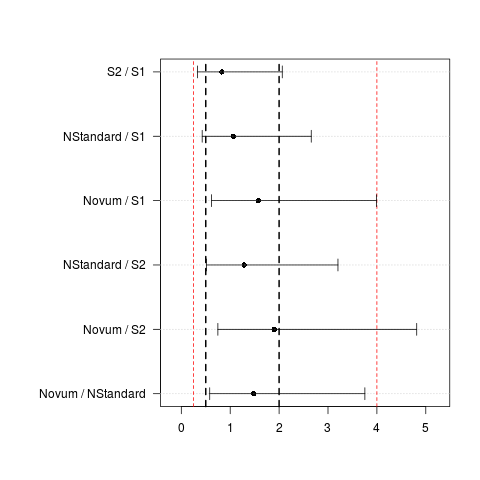
plotCI(UnlogCI(CIraw), lines=c(0.25,0.5,2,4),
lineslwd=c(1,2,2,1), linescol=c("red","black","black","red"))
# # # # # # #
# # # CI for ratios of means
# # # for models on the log-link
data(Diptera)
# Larval mortality:
fit2<-glm(Ges~Treatment, data=Diptera, family=quasipoisson)
anova(fit2, test="F")
library(multcomp)
comp<-glht(fit2, mcp(Treatment="Tukey"))
CIadj<-CIGLM(comp,method="Adj")
CIadj
UnlogCI(CIadj)
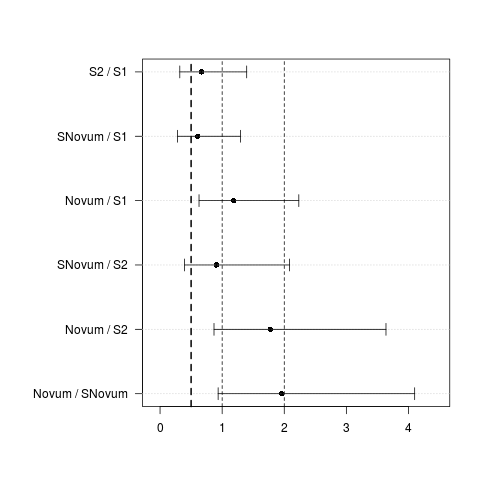
plotCI(UnlogCI(CIadj), lines=c(0.5,1,2), lineslwd=c(2,1,1))
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(BSagri)
Loading required package: gamlss
Loading required package: splines
Loading required package: gamlss.data
Loading required package: gamlss.dist
Loading required package: MASS
Loading required package: nlme
Loading required package: parallel
********** GAMLSS Version 4.4-0 **********
For more on GAMLSS look at http://www.gamlss.org/
Type gamlssNews() to see new features/changes/bug fixes.
Loading required package: multcomp
Loading required package: mvtnorm
Loading required package: survival
Attaching package: 'survival'
The following object is masked from 'package:gamlss.data':
leukemia
Loading required package: TH.data
Attaching package: 'TH.data'
The following object is masked from 'package:MASS':
geyser
Loading required package: MCPAN
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/BSagri/UnlogCI.Rd_%03d_medium.png", width=480, height=480)
> ### Name: UnlogCI
> ### Title: Transform confidence intervals from glm fits.
> ### Aliases: UnlogCI UnlogCI.glht
> ### Keywords: htest
>
> ### ** Examples
>
>
> # # # CI for odds ratios
> # # # for models on the logit-link
>
> data(Feeding)
>
> # Larval mortality:
>
> Feeding$Lmort <- Feeding$Total - Feeding$Pupating
>
> fit1<-glm(cbind(Pupating,Lmort)~Variety,data=Feeding, family=quasibinomial)
> anova(fit1, test="F")
Analysis of Deviance Table
Model: quasibinomial, link: logit
Response: cbind(Pupating, Lmort)
Terms added sequentially (first to last)
Df Deviance Resid. Df Resid. Dev F Pr(>F)
NULL 31 59.623
Variety 3 3.3623 28 56.260 0.6478 0.5909
>
> library(multcomp)
>
> comp<-glht(fit1, mcp(Variety="Tukey"))
>
> CIraw<-CIGLM(comp,method="Raw")
>
> CIraw
Simultaneous Confidence Intervals
Multiple Comparisons of Means: Tukey Contrasts
Fit: glm(formula = cbind(Pupating, Lmort) ~ Variety, family = quasibinomial,
data = Feeding)
Quantile = 1.96
95% confidence level
Linear Hypotheses:
Estimate lwr upr
S2 - S1 == 0 -0.18768 -1.10029 0.72493
NStandard - S1 == 0 0.06289 -0.85152 0.97729
Novum - S1 == 0 0.45291 -0.47882 1.38465
NStandard - S2 == 0 0.25057 -0.66316 1.16431
Novum - S2 == 0 0.64060 -0.29048 1.57168
Novum - NStandard == 0 0.39003 -0.54281 1.32287
>
> UnlogCI(CIraw)
glm(formula = cbind(Pupating, Lmort) ~ Variety, family = quasibinomial,
data = Feeding)
Confidence intervals for odds ratios
with the odds defined as p(Pupating)/(1-p(Pupating))
Estimate lwr upr
S2 / S1 0.829 0.333 2.065
NStandard / S1 1.065 0.427 2.657
Novum / S1 1.573 0.620 3.993
NStandard / S2 1.285 0.515 3.204
Novum / S2 1.898 0.748 4.815
Novum / NStandard 1.477 0.581 3.754
Estimated quantile = 1.96
>
> plotCI(UnlogCI(CIraw), lines=c(0.25,0.5,2,4),
+ lineslwd=c(1,2,2,1), linescol=c("red","black","black","red"))
>
>
> # # # # # # #
>
> # # # CI for ratios of means
> # # # for models on the log-link
>
> data(Diptera)
>
> # Larval mortality:
>
> fit2<-glm(Ges~Treatment, data=Diptera, family=quasipoisson)
> anova(fit2, test="F")
Analysis of Deviance Table
Model: quasipoisson, link: log
Response: Ges
Terms added sequentially (first to last)
Df Deviance Resid. Df Resid. Dev F Pr(>F)
NULL 31 621.12
Treatment 3 145.62 28 475.50 2.6403 0.06892 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
>
> library(multcomp)
>
> comp<-glht(fit2, mcp(Treatment="Tukey"))
>
> CIadj<-CIGLM(comp,method="Adj")
>
> CIadj
Simultaneous Confidence Intervals
Multiple Comparisons of Means: Tukey Contrasts
Fit: glm(formula = Ges ~ Treatment, family = quasipoisson, data = Diptera)
Quantile = 2.5664
95% family-wise confidence level
Linear Hypotheses:
Estimate lwr upr
S2 - S1 == 0 -0.40637 -1.14578 0.33305
SNovum - S1 == 0 -0.50303 -1.26464 0.25857
Novum - S1 == 0 0.16900 -0.46596 0.80395
SNovum - S2 == 0 -0.09667 -0.92710 0.73377
Novum - S2 == 0 0.57536 -0.14070 1.29143
Novum - SNovum == 0 0.67203 -0.06693 1.41099
>
> UnlogCI(CIadj)
glm(formula = Ges ~ Treatment, family = quasipoisson, data = Diptera)
Confidence intervals for the ratios of abundance
Estimate lwr upr
S2 / S1 0.666 0.318 1.395
SNovum / S1 0.605 0.282 1.295
Novum / S1 1.184 0.628 2.234
SNovum / S2 0.908 0.396 2.083
Novum / S2 1.778 0.869 3.638
Novum / SNovum 1.958 0.935 4.100
Estimated quantile = 2.5664
>
> plotCI(UnlogCI(CIadj), lines=c(0.5,1,2), lineslwd=c(2,1,1))
>
>
>
>
>
>
>
> dev.off()
null device
1
>
|
|
 providing
providing  .
.