Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Finds Synteny in a Sequence DatabaseDescriptionFinds syntenic blocks between groups of sequences in a database. Usage
FindSynteny(dbFile,
tblName = "Seqs",
identifier = "",
useFrames = FALSE,
alphabet = c("MF", "ILV", "A", "C", "WYQHP", "G", "TSN", "RK", "DE"),
geneticCode = GENETIC_CODE,
sepCost = -0.01,
gapCost = -0.2,
shiftCost = -20,
codingCost = -3,
maxSep = 5000,
maxGap = 5000,
minScore = 200,
dropScore = -100,
maskRepeats = TRUE,
storage = 0.5,
processors = 1,
verbose = TRUE)
Arguments
DetailsLong nucleotide sequences, such as genomes, are often not collinear, or may be composed of many smaller segments (e.g., contigs). ValueAn object of class “Synteny”. Author(s)Erik Wright DECIPHER@cae.wisc.edu See Also
Examples
db <- system.file("extdata", "Influenza.sqlite", package="DECIPHER")
synteny <- FindSynteny(db, useFrames=TRUE, minScore=50)
synteny
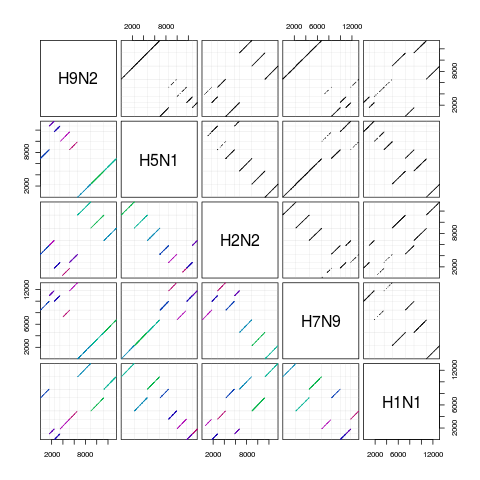
pairs(synteny) # scatterplot matrix
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(DECIPHER)
Loading required package: Biostrings
Loading required package: BiocGenerics
Loading required package: parallel
Attaching package: 'BiocGenerics'
The following objects are masked from 'package:parallel':
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLB
The following objects are masked from 'package:stats':
IQR, mad, xtabs
The following objects are masked from 'package:base':
Filter, Find, Map, Position, Reduce, anyDuplicated, append,
as.data.frame, cbind, colnames, do.call, duplicated, eval, evalq,
get, grep, grepl, intersect, is.unsorted, lapply, lengths, mapply,
match, mget, order, paste, pmax, pmax.int, pmin, pmin.int, rank,
rbind, rownames, sapply, setdiff, sort, table, tapply, union,
unique, unsplit
Loading required package: S4Vectors
Loading required package: stats4
Attaching package: 'S4Vectors'
The following objects are masked from 'package:base':
colMeans, colSums, expand.grid, rowMeans, rowSums
Loading required package: IRanges
Loading required package: XVector
Loading required package: RSQLite
Loading required package: DBI
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/DECIPHER/FindSynteny.Rd_%03d_medium.png", width=480, height=480)
> ### Name: FindSynteny
> ### Title: Finds Synteny in a Sequence Database
> ### Aliases: FindSynteny
>
> ### ** Examples
>
> db <- system.file("extdata", "Influenza.sqlite", package="DECIPHER")
> synteny <- FindSynteny(db, useFrames=TRUE, minScore=50)
| | | 0% | |==== | 5% | |======= | 10% | |========== | 15% | |============== | 20% | |================== | 25% | |===================== | 30% | |======================== | 35% | |============================ | 40% | |================================ | 45% | |=================================== | 50% | |====================================== | 55% | |========================================== | 60% | |============================================== | 65% | |================================================= | 70% | |==================================================== | 75% | |======================================================== | 80% | |============================================================ | 85% | |=============================================================== | 90% | |================================================================== | 95% | |======================================================================| 100%
Time difference of 1.12 secs
> synteny
H9N2 H5N1 H2N2 H7N9 H1N1
H9N2 8 seqs 67% hits 76% hits 65% hits 70% hits
H5N1 8 blocks 8 seqs 68% hits 65% hits 78% hits
H2N2 8 blocks 8 blocks 8 seqs 73% hits 68% hits
H7N9 8 blocks 8 blocks 8 blocks 8 seqs 71% hits
H1N1 8 blocks 8 blocks 8 blocks 8 blocks 8 seqs
> pairs(synteny) # scatterplot matrix
>
>
>
>
>
> dev.off()
null device
1
>
|