Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
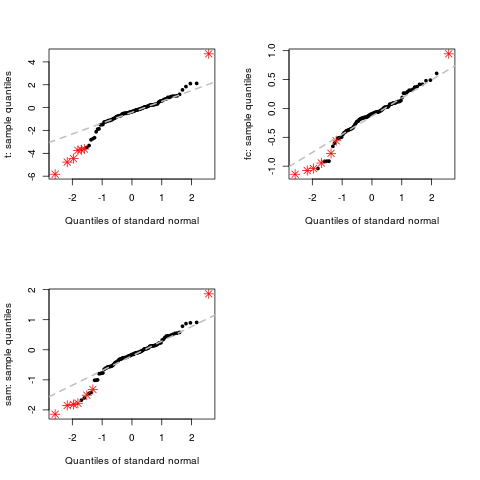
Normal Q-Q Plot for DEDS ObjectsDescriptionThe function Usage## S3 method for class 'DEDS' qqnorm(y, subset=c(1:nrow(y$stats)), xlab = "Quantiles of standard normal", thresh = 0.05, col = palette(), pch, ...) Arguments
DetailsThe function For DEDS objects that are created by the function
Author(s)Yuanyuan Xiao, yxiao@itsa.ucsf.edu, See Also
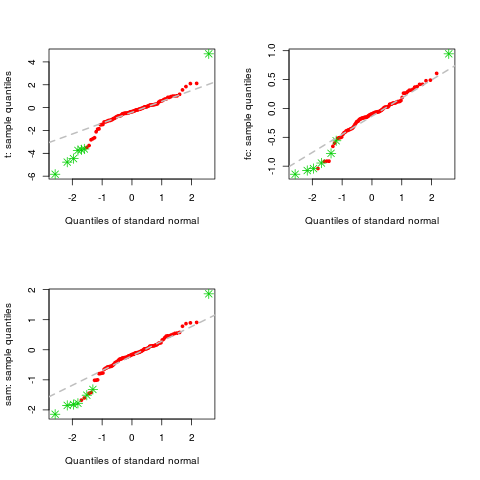
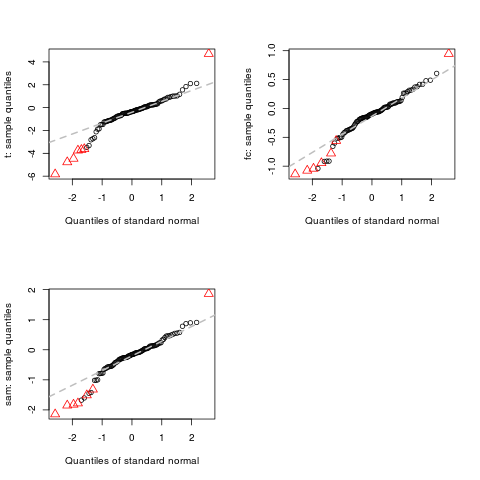
ExamplesX <- matrix(rnorm(1000,0,0.5), nc=10) L <- rep(0:1,c(5,5)) # genes 1-10 are differentially expressed X[1:10,6:10]<-X[1:10,6:10]+1 # DEDS summarizing t, fc and sam d <- deds.stat.linkC(X, L, B=200) # qqnorm for t, fc and sam qqnorm(d) # change points color qqnorm(d, col=c(2,3)) # change points type qqnorm(d, pch=c(1,2)) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(DEDS)
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/DEDS/qqnorm.DEDS.Rd_%03d_medium.png", width=480, height=480)
> ### Name: qqnorm-methods
> ### Title: Normal Q-Q Plot for DEDS Objects
> ### Aliases: qqnorm.DEDS qqnorm-methods qqnorm
> ### Keywords: hplot
>
> ### ** Examples
>
> X <- matrix(rnorm(1000,0,0.5), nc=10)
> L <- rep(0:1,c(5,5))
>
> # genes 1-10 are differentially expressed
> X[1:10,6:10]<-X[1:10,6:10]+1
> # DEDS summarizing t, fc and sam
> d <- deds.stat.linkC(X, L, B=200)
We'll do random permutations, B = 200
Two-sample Statistics:
t FC SAM
E of the orginial data is: 5.828 1.140 2.143
b=2 b=4 b=6 b=8 b=10 b=12 b=14 b=16 b=18 b=20
b=22 b=24 b=26 b=28 b=30 b=32 b=34 b=36 b=38 b=40
b=42 b=44 b=46 b=48 b=50 b=52 b=54 b=56 b=58 b=60
b=62 b=64 b=66 b=68 b=70 b=72 b=74 b=76 b=78 b=80
b=82 b=84 b=86 b=88 b=90 b=92 b=94 b=96 b=98 b=100
b=102 b=104 b=106 b=108 b=110 b=112 b=114 b=116 b=118 b=120
b=122 b=124 b=126 b=128 b=130 b=132 b=134 b=136 b=138 b=140
b=142 b=144 b=146 b=148 b=150 b=152 b=154 b=156 b=158 b=160
b=162 b=164 b=166 b=168 b=170 b=172 b=174 b=176 b=178 b=180
b=182 b=184 b=186 b=188 b=190 b=192 b=194 b=196 b=198 b=200
After permutation , E is set at: 8.922 1.284 2.479
Summarizing DEDS results for 200 permutations and 100 genes, please wait...
>
> # qqnorm for t, fc and sam
> qqnorm(d)
> # change points color
> qqnorm(d, col=c(2,3))
> # change points type
> qqnorm(d, pch=c(1,2))
>
>
>
>
>
> dev.off()
null device
1
>
|