Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Designs based on Strauss processDescriptionSpace-Filling Designs based on Strauss process UsagestraussDesign(n,dimension,RND,alpha=0.5,repulsion=0.001,NMC=1000,
constraints1D=0,repulsion1D=0.0001,seed=NULL)
Arguments
DetailsStrauss designs are Space-Filling designs initially defined from Strauss process: π (X) = k gamma^s(X) where s(X) is is the number of pairs of points (xi,xj) of the design X = ( x1, ..., xn
ight) that are separated by a distance no greater than the radius of interaction For the general case, a stochastic simulation is used to construct a Markov chain which converges to a spatial density of points π(X) described by the Strauss-Gibbs potential. In practice, the Metropolis-Hastings algorithm is implemented to simulate a distribution of points which converges to the stationary law: π(X) = k exp(-U(X)) with a potentiel U defined by: beta Sum_{i<j} phi(|| xi-xj ||) where beta = -ln(gamma), phi (h) = (1-h/RND)^{alpha} if h <= The input parameters of - - - - ValueA list containing:
Author(s)J. Franco ReferencesJ. Franco, X. Bay, B. Corre and D. Dupuy (2008) Planification d experiences numeriques a partir du processus ponctuel de Strauss http://hal.archives-ouvertes.fr/hal-00260701/fr/ (March 06,2008). Examples
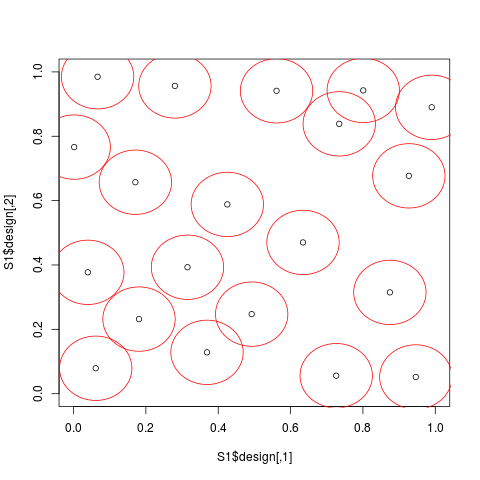
# Strauss-Gibbs designs in dimension 2 (n=20 points)
S1 <- straussDesign(n=20,dimension=2,RND=0.2)
plot(S1$design,xlim=c(0,1),ylim=c(0,1))
theta <- seq(0,2*pi,by =2*pi/(100 - 1))
for(i in 1:S1$n){
lines(S1$design[i,1]+S1$radius/2*cos(theta),
S1$design[i,2]+S1$radius/2*sin(theta),col='red')
}
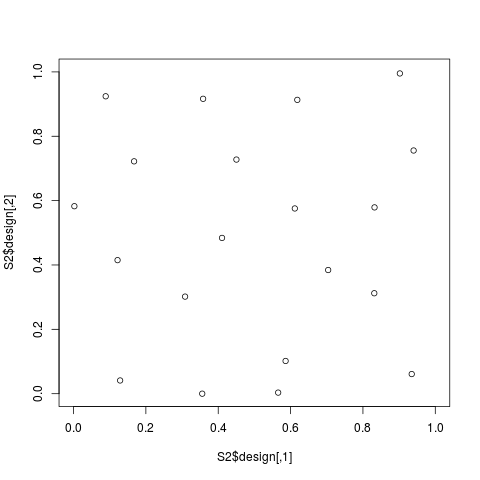
# 2D-Strauss design
S2 <- straussDesign(n=20,dimension=2,RND=0.2,NMC=200,
constraints1D=0,alpha=0,repulsion=0.01)
plot(S2$design,xlim=c(0,1),ylim=c(0,1))
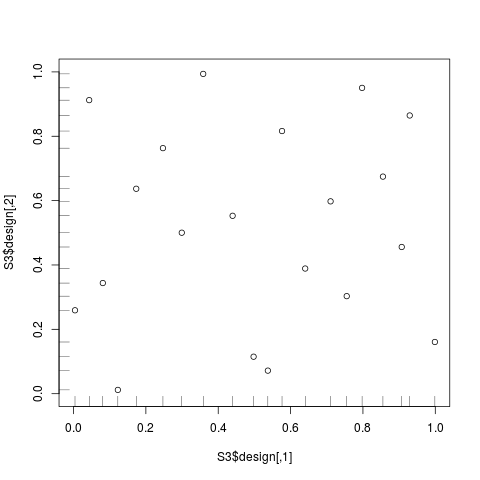
# 2D-Strauss designs with constraints on the axis
S3 <- straussDesign(n=20,dimension=2,RND=0.18,NMC=200,
constraints1D=1,alpha=0.5,repulsion=0.1,repulsion1D=0.01)
plot(S3$design,xlim=c(0,1),ylim=c(0,1))
rug(S3$design[,1],side=1)
rug(S3$design[,2],side=2)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(DiceDesign)
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/DiceDesign/straussDesign.Rd_%03d_medium.png", width=480, height=480)
> ### Name: straussDesign
> ### Title: Designs based on Strauss process
> ### Aliases: straussDesign
> ### Keywords: design
>
> ### ** Examples
>
> # Strauss-Gibbs designs in dimension 2 (n=20 points)
> S1 <- straussDesign(n=20,dimension=2,RND=0.2)
> plot(S1$design,xlim=c(0,1),ylim=c(0,1))
> theta <- seq(0,2*pi,by =2*pi/(100 - 1))
> for(i in 1:S1$n){
+ lines(S1$design[i,1]+S1$radius/2*cos(theta),
+ S1$design[i,2]+S1$radius/2*sin(theta),col='red')
+ }
> # 2D-Strauss design
> S2 <- straussDesign(n=20,dimension=2,RND=0.2,NMC=200,
+ constraints1D=0,alpha=0,repulsion=0.01)
>
> plot(S2$design,xlim=c(0,1),ylim=c(0,1))
>
> # 2D-Strauss designs with constraints on the axis
> S3 <- straussDesign(n=20,dimension=2,RND=0.18,NMC=200,
+ constraints1D=1,alpha=0.5,repulsion=0.1,repulsion1D=0.01)
>
> plot(S3$design,xlim=c(0,1),ylim=c(0,1))
> rug(S3$design[,1],side=1)
> rug(S3$design[,2],side=2)
>
>
>
>
>
> dev.off()
null device
1
>
|