Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Generate the Transition Density of a Bivariate Generalized Quadratic Diffusion Model (2D GQD).Description
where
and
Usage
BiGQD.density(Xs, Ys, Xt, Yt, s, t, delt=1/100, Dtype='Saddlepoint', print.output=TRUE,
eval.density=TRUE)
Arguments
Details
Value
WarningWarning [1]: The system of ODEs that dictate the evolution of the cumulants do so approximately. Thus, although it is unlikely such cases will be encountered in inferential contexts, it is worth checking (by simulation) whether cumulants accurately replicate those of the target GQD. Furthermore, it may in some cases occur that the cumulants are indeed accurate whilst the density approximation fails. This can again be verified by simulation. Warning [2]:
The parameter Author(s)Etienne A.D. Pienaar: etiannead@gmail.com ReferencesUpdates available on GitHub at https://github.com/eta21. Daniels, H.E. 1954 Saddlepoint approximations in statistics. Ann. Math. Stat., 25:631–650. Eddelbuettel, D. and Romain, F. 2011 Rcpp: Seamless R and C++ integration. Journal of Statistical Software, 40(8):1–18,. URL http://www.jstatsoft.org/v40/i08/. Eddelbuettel, D. 2013 Seamless R and C++ Integration with Rcpp. New York: Springer. ISBN 978-1-4614-6867-7. Eddelbuettel, D. and Sanderson, C. 2014 Rcpparmadillo: Accelerating R with high-performance C++ linear algebra. Computational Statistics and Data Analysis, 71:1054–1063. URL http://dx.doi.org/10.1016/j.csda.2013.02.005. Feagin, T. 2007 A tenth-order Runge-Kutta method with error estimate. In Proceedings of the IAENG Conf. on Scientifc Computing. Varughese, M.M. 2013 Parameter estimation for multivariate diffusion systems. Comput. Stat. Data An., 57:417–428. See AlsoSee Examples
#===============================================================================
# Generate the transition density of a stochastic perturbed Lotka-Volterra
# preditor-prey model, with state-dependent volatility:
# dX = (1.5X-0.4*X*Y)dt +sqrt(0.05*X)dWt
# dY = (-1.5Y+0.4*X*Y-0.2*Y^2)dt +sqrt(0.10*Y)dBt
#-------------------------------------------------------------------------------
# Remove any existing coefficients
GQD.remove()
# Define the X dimesnion coefficients
a10 = function(t){1.5}
a11 = function(t){-0.4}
c10 = function(t){0.05}
# Define the Y dimension coefficients
b01 = function(t){-1.5}
b11 = function(t){0.4}
b02 = function(t){-0.2}
f01 = function(t){0.1}
# Approximate the transition density
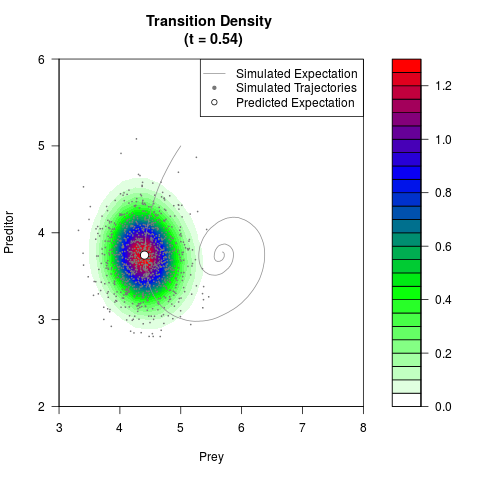
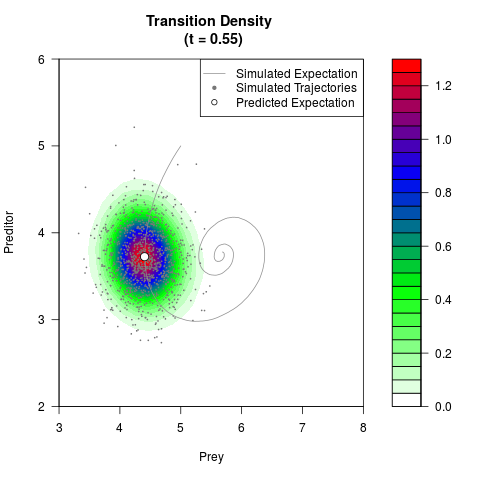
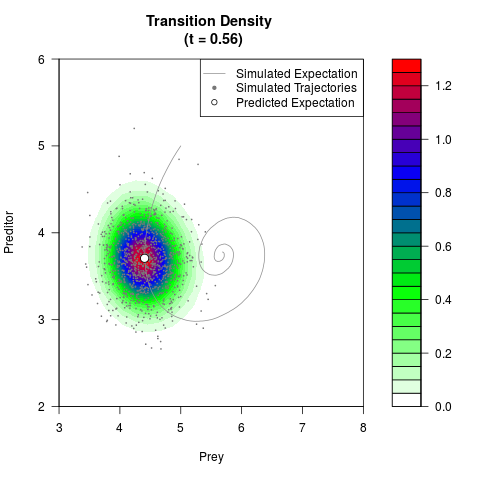
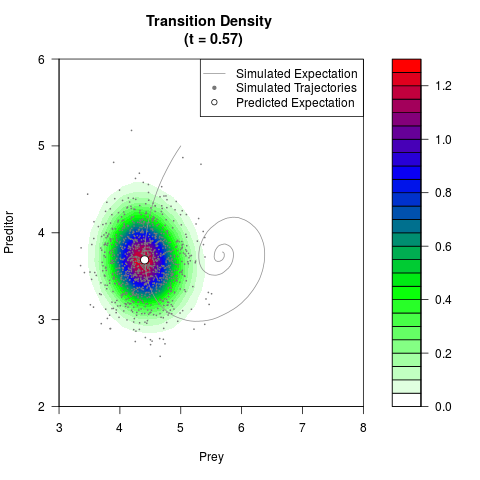
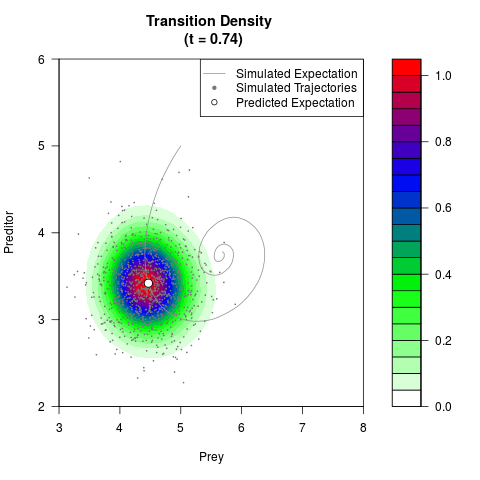
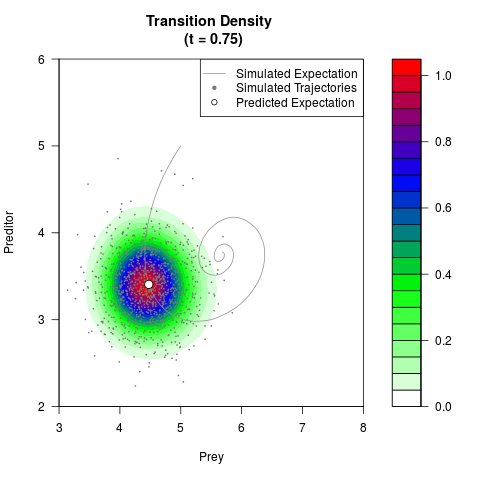
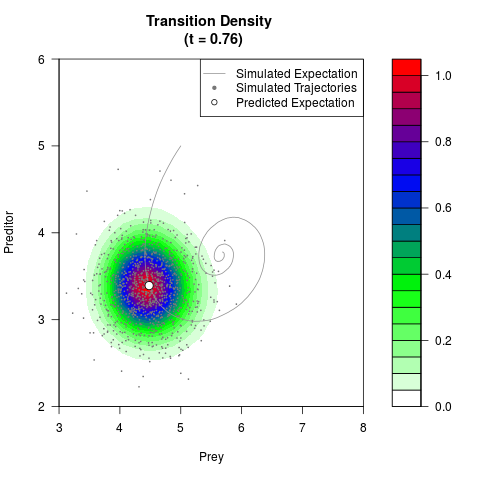
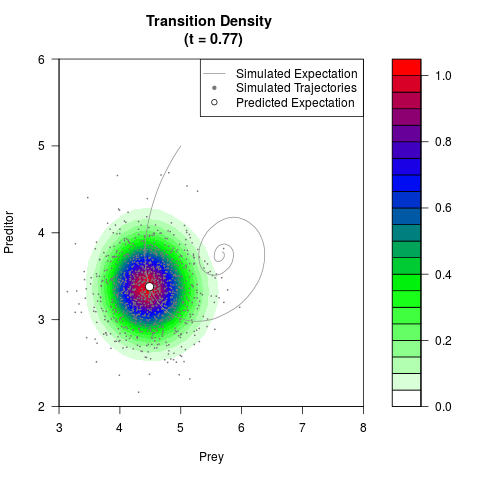
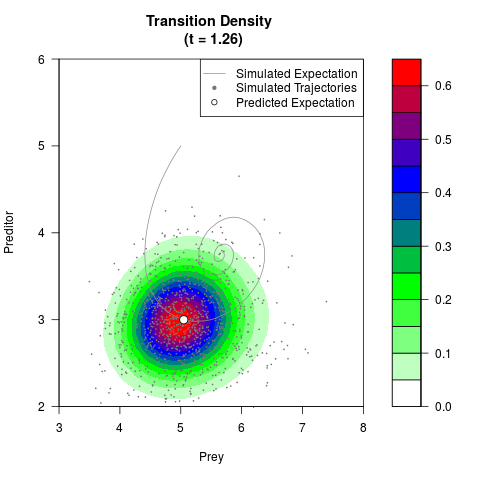
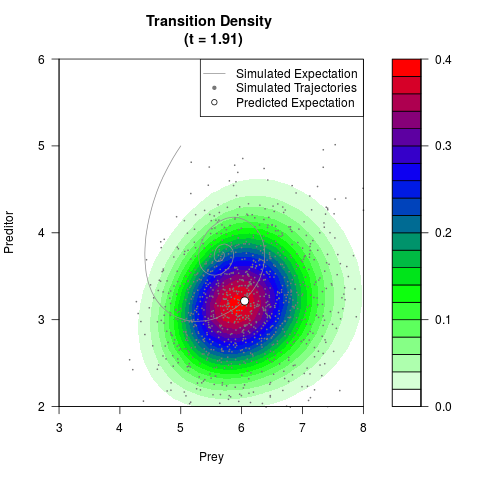
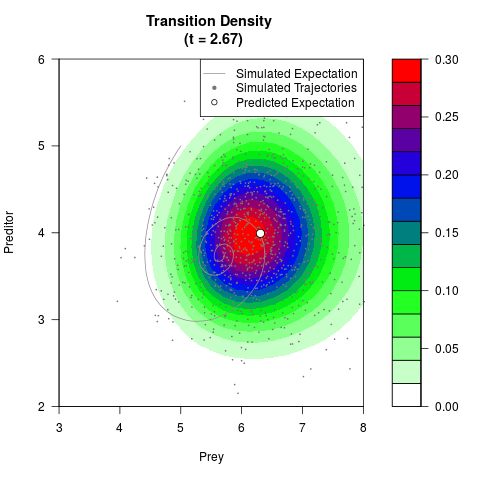
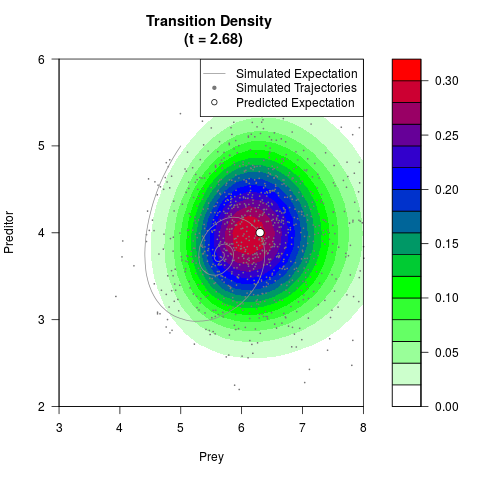
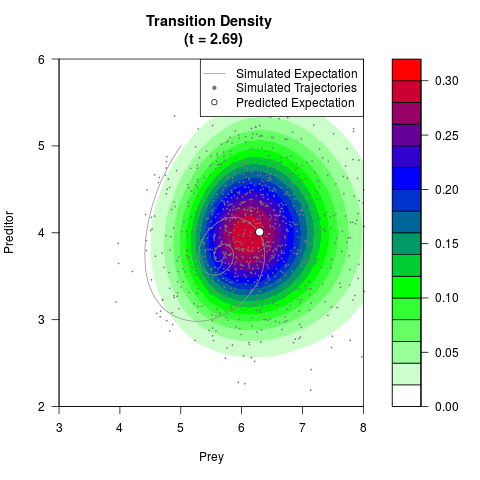
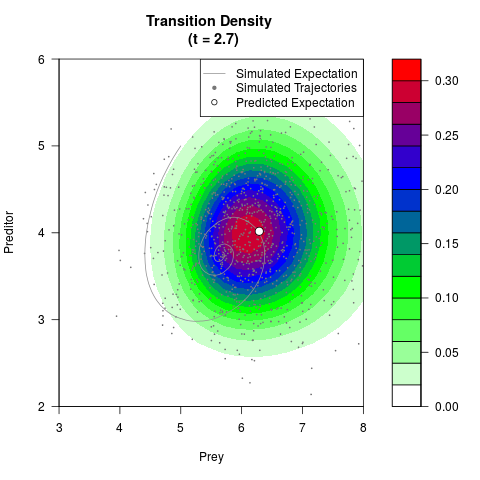
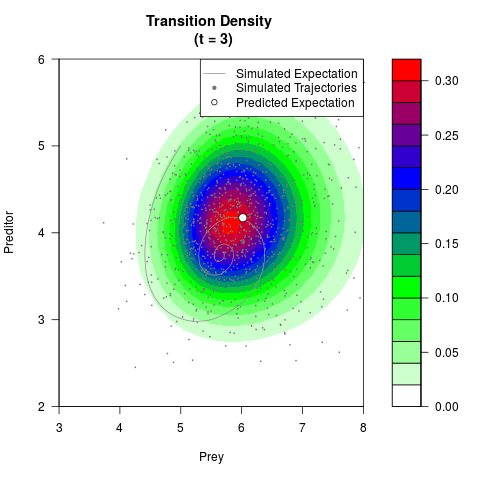
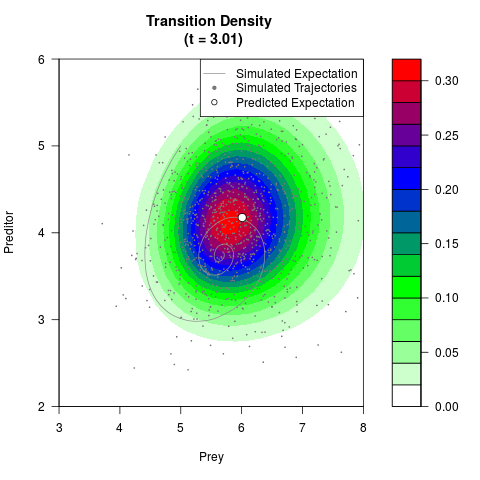
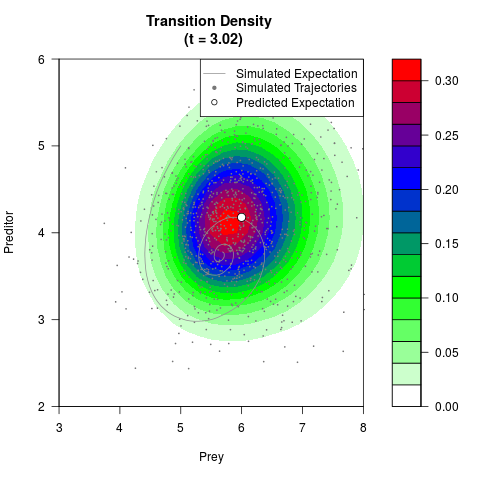
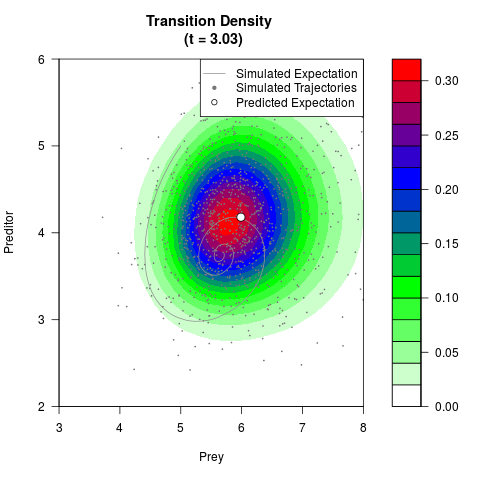
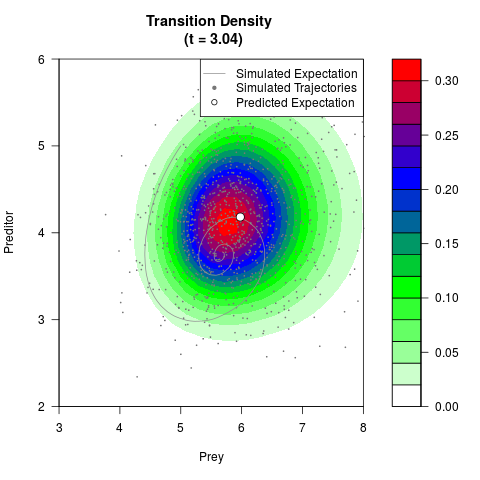
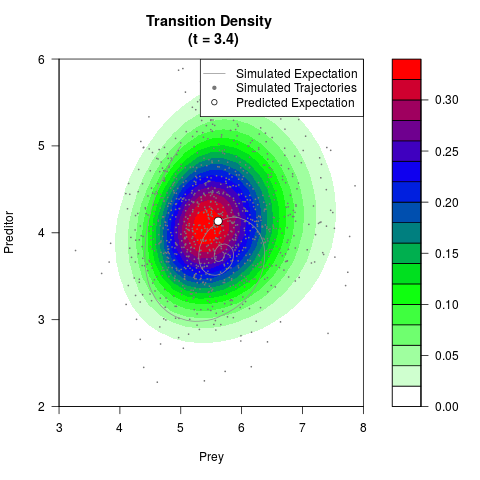
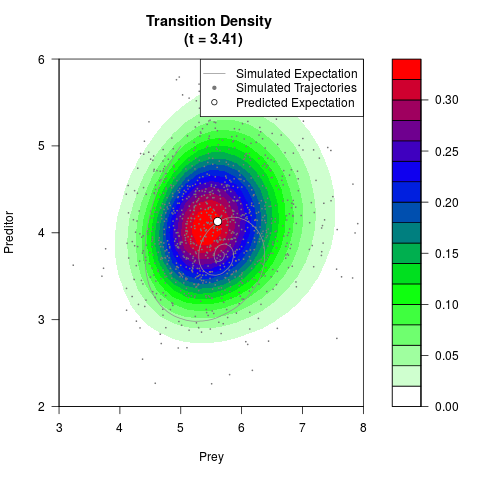
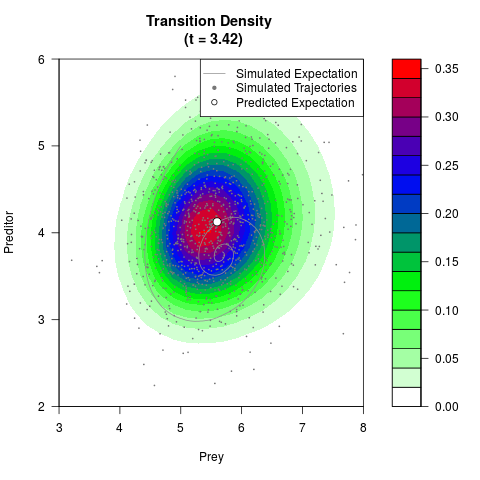
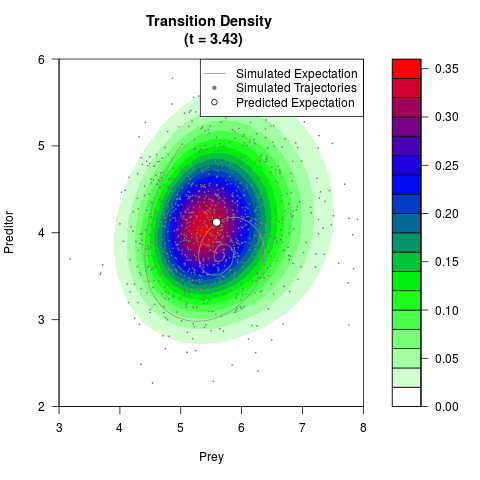
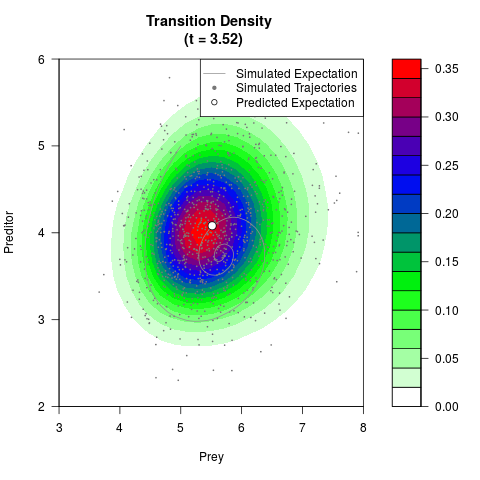
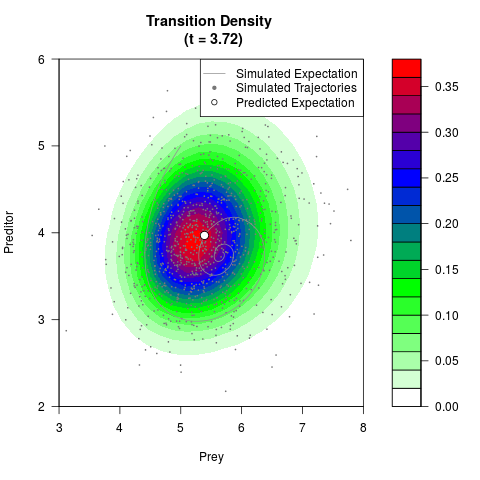
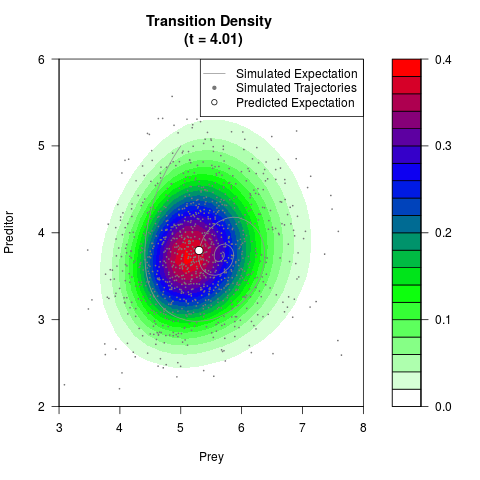
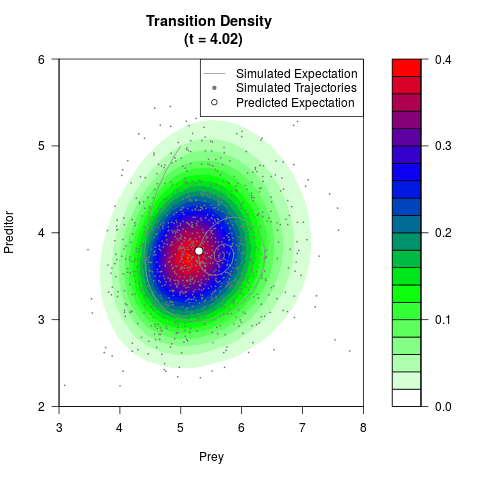
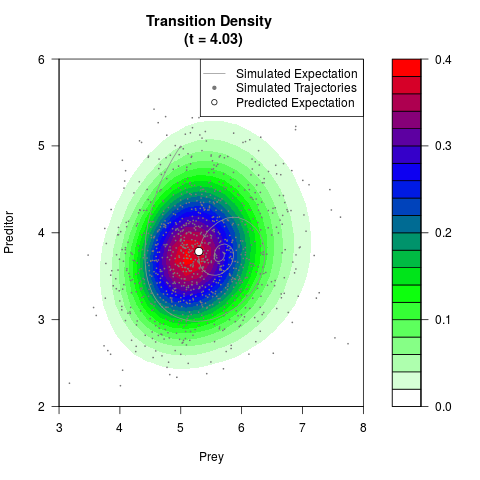
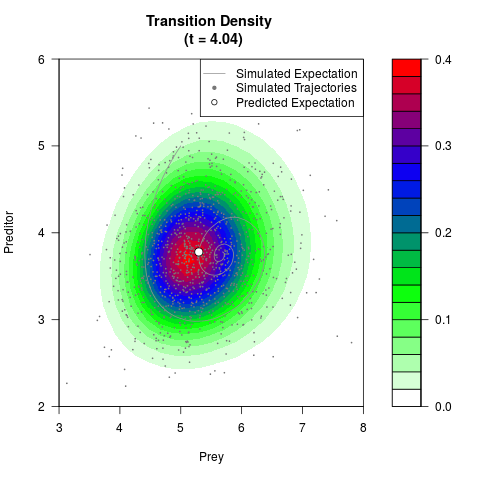
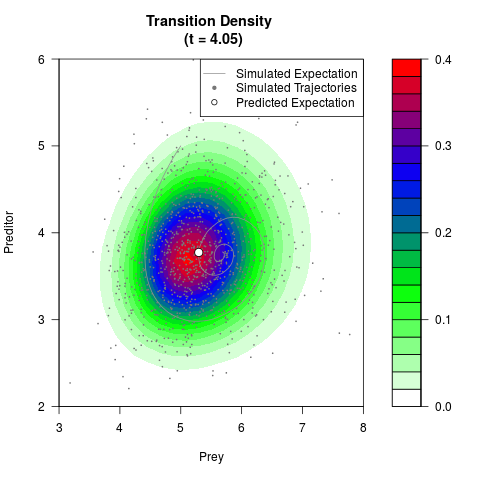
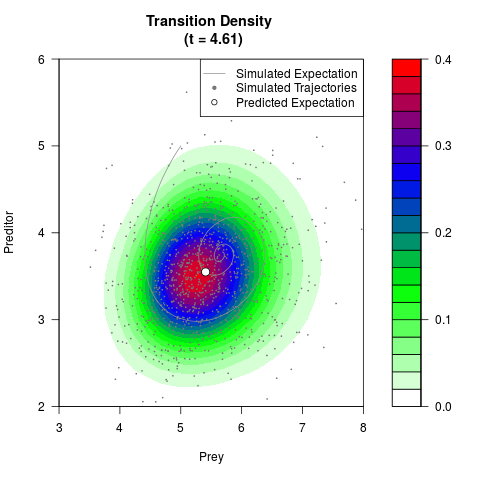
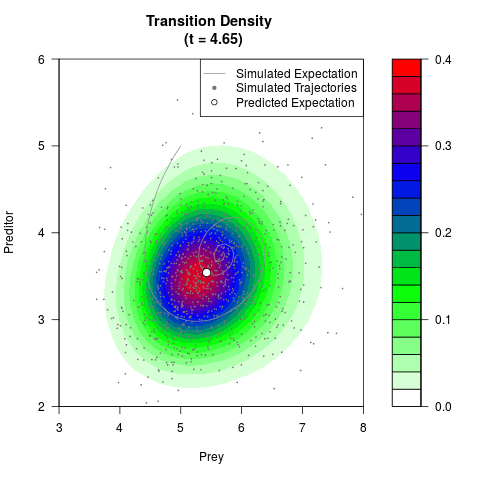
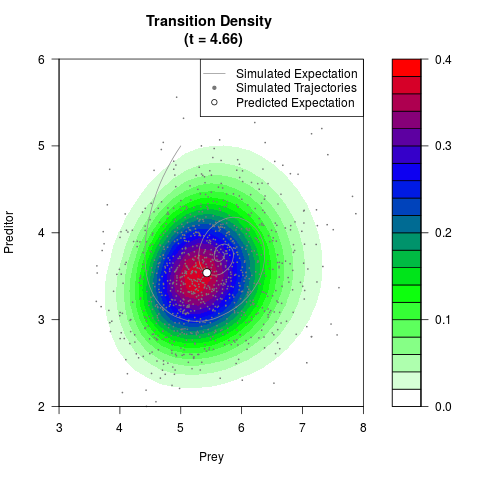
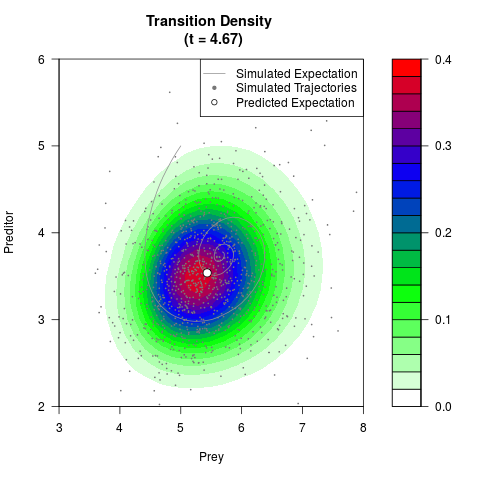
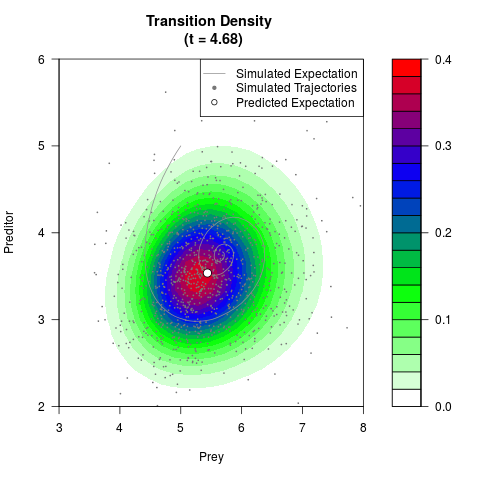
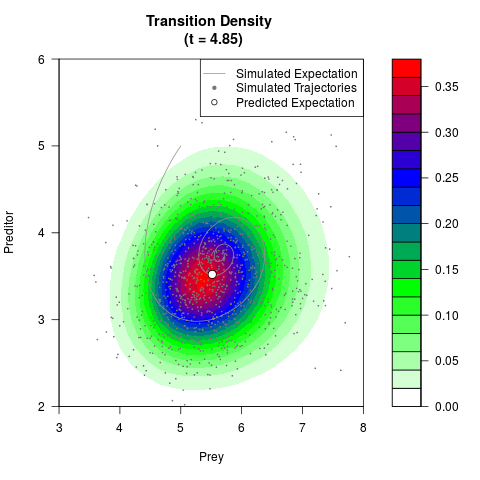
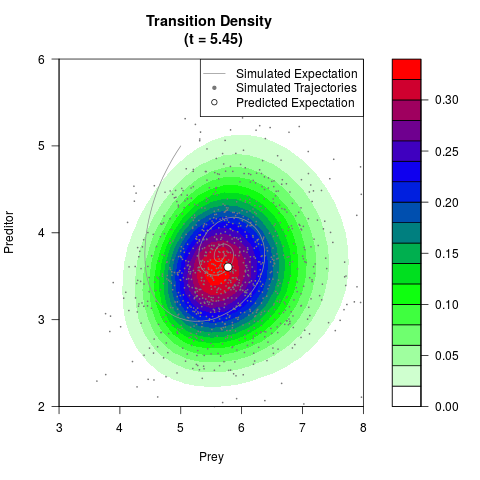
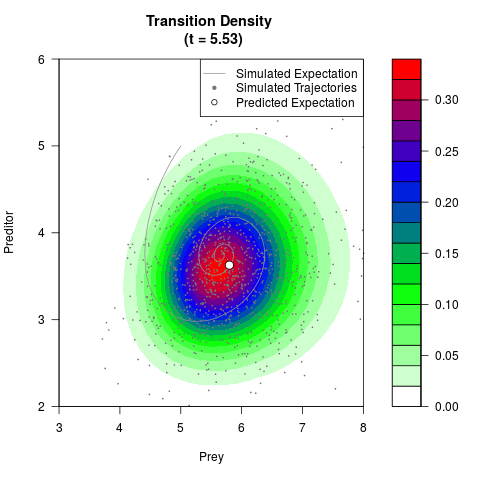
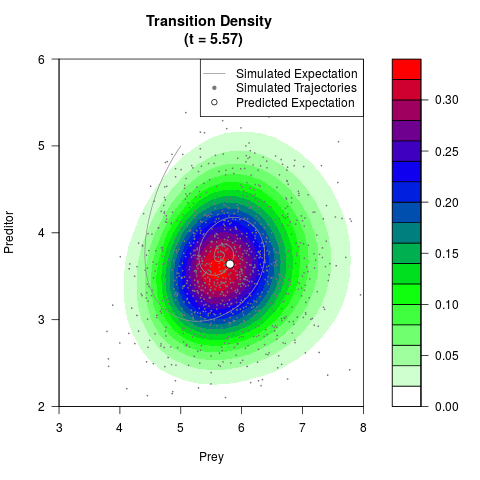
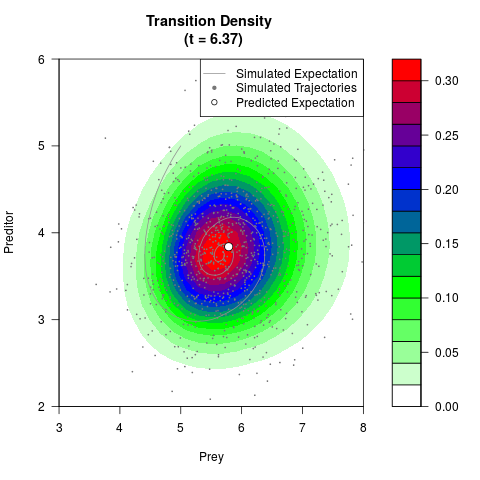
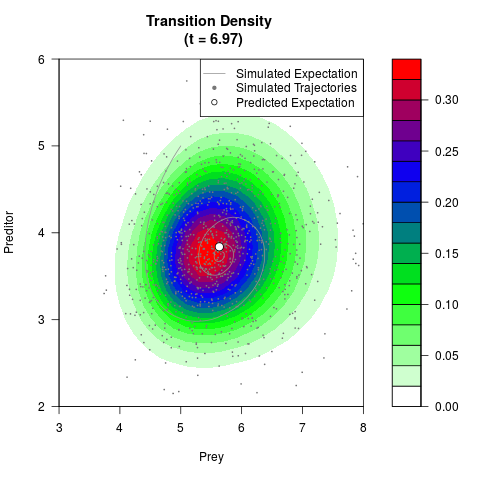
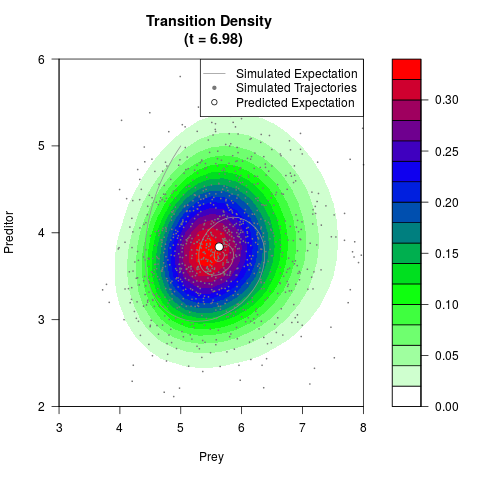
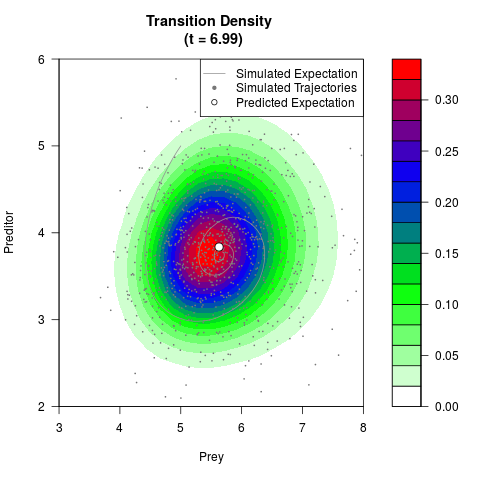
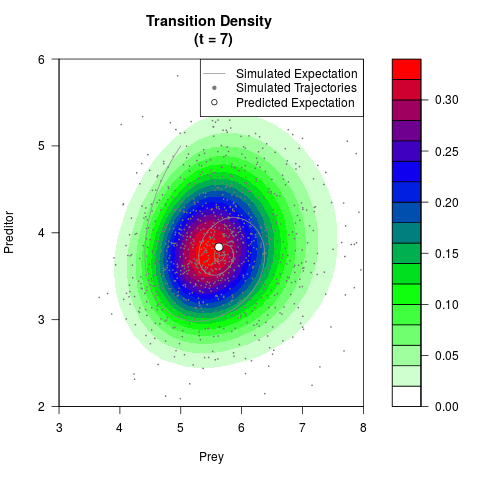
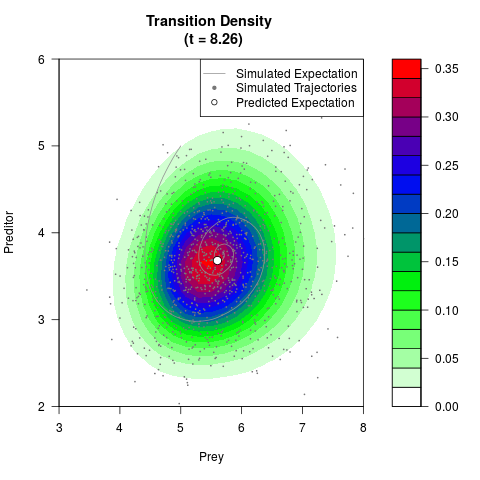
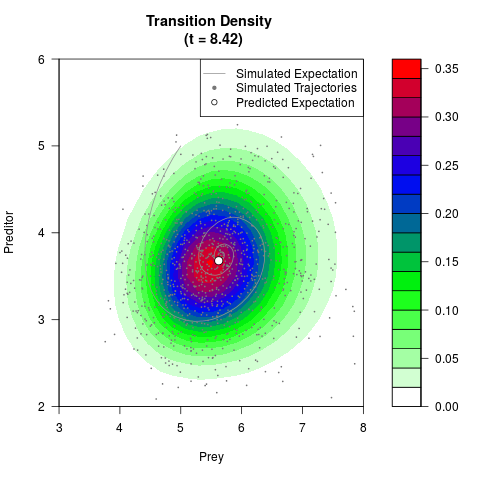
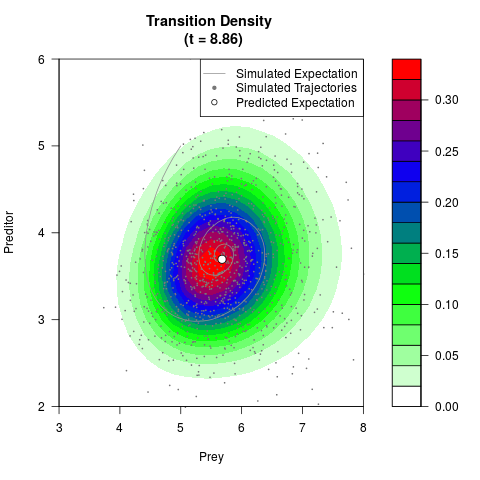
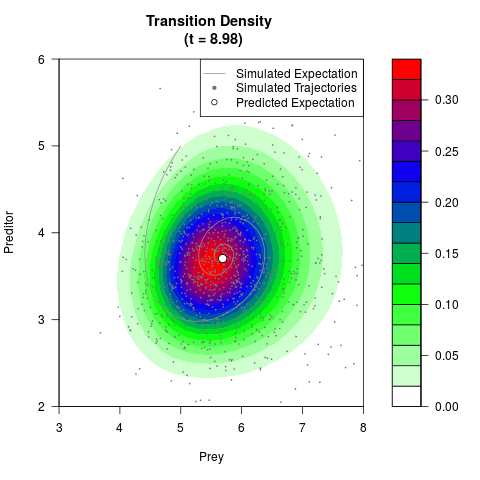
res = BiGQD.density(5,5,seq(3,8,length=25),seq(2,6,length=25),0,10,1/100)
#------------------------------------------------------------------------------
# Visuallize the density
#------------------------------------------------------------------------------
par(ask=FALSE)
# Load simulated trajectory of the joint expectation:
data(SDEsim3)
attach(SDEsim3)
# We will simulate some trajectories (crudely) as well:
N=1000; delt= 1/100 # 1000 trajectories
X1=rep(5,N) # Initial values for each trajectory
X2=rep(5,N)
for(i in 1:1001)
{
# Applly Euler-Murayama scheme to the LV-model
X1=pmax(X1+(a10(d)*X1+a11(d)*X1*X2)*delt+sqrt(c10(d)*X1)*rnorm(N,sd=sqrt(delt)),0)
X2=pmax(X2+(b01(d)*X2+b11(d)*X1*X2+b02(d)*X2^2)*delt+sqrt(f01(d)*X2)*rnorm(N,sd=sqrt(delt)),0)
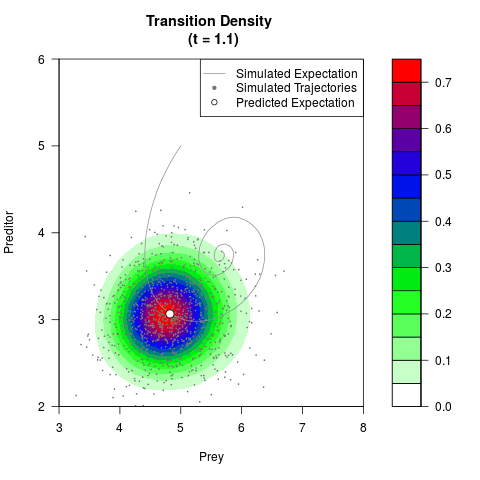
# Now illustrate the density:
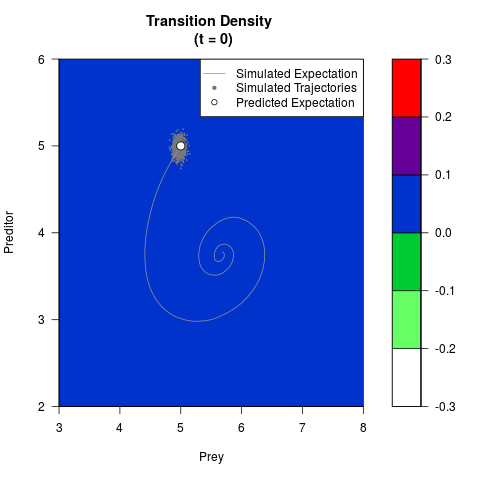
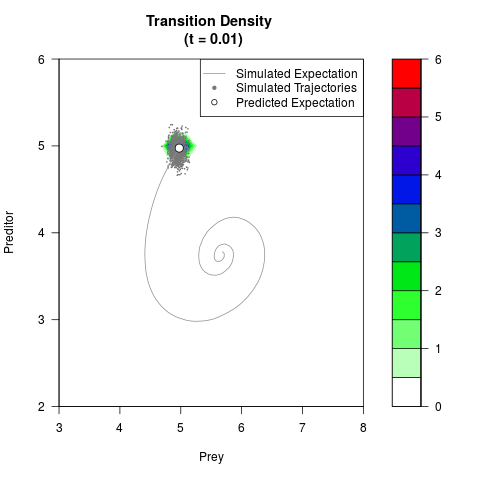
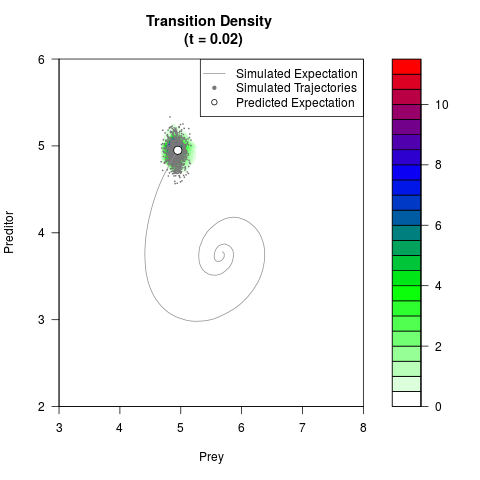
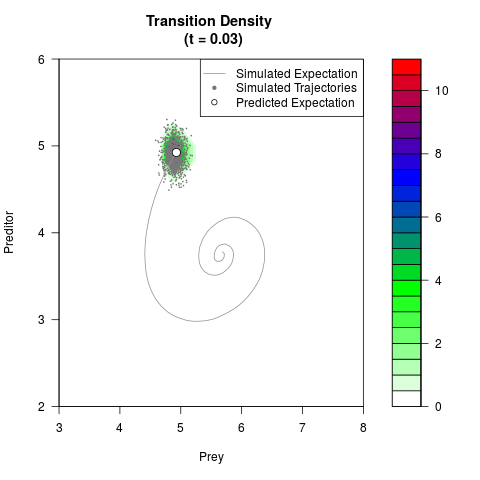
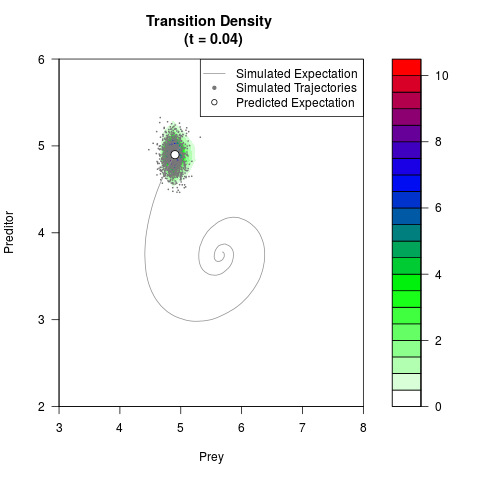
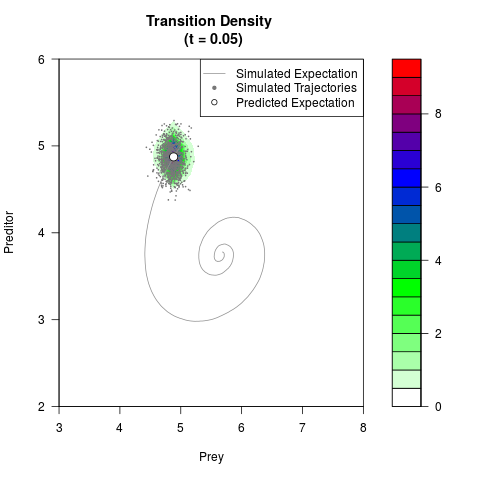
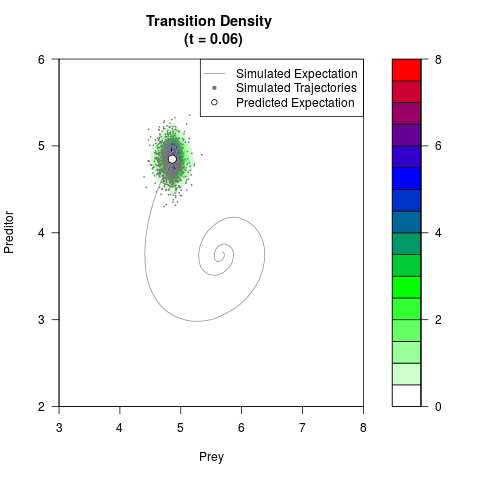
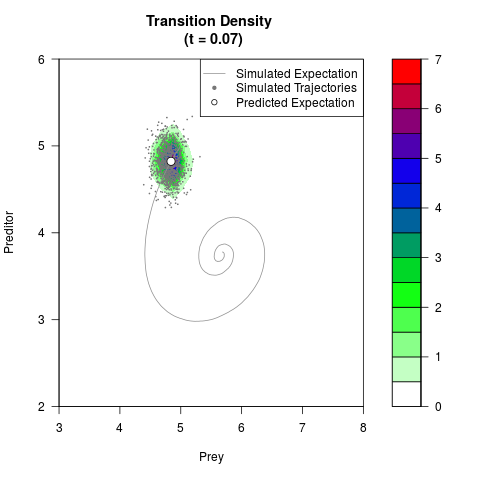
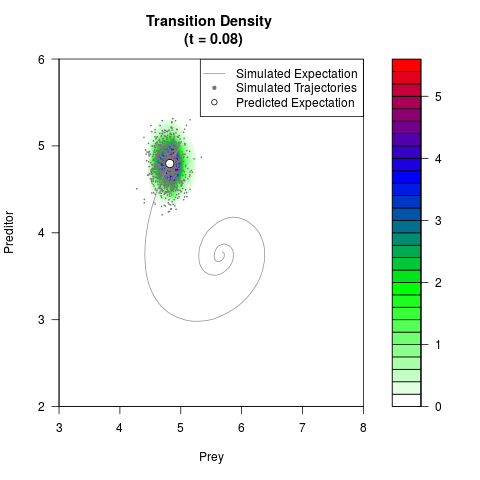
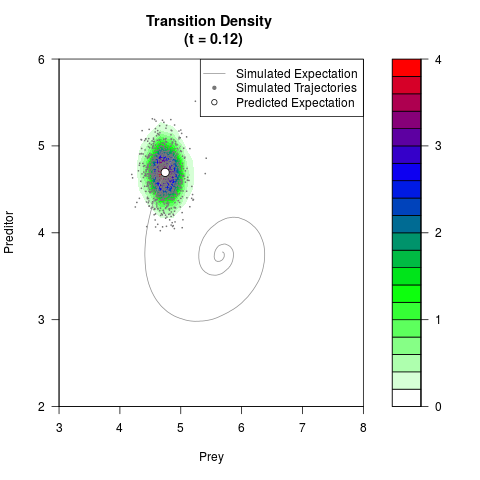
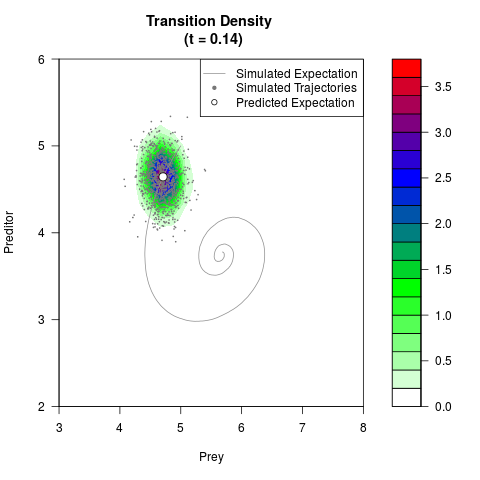
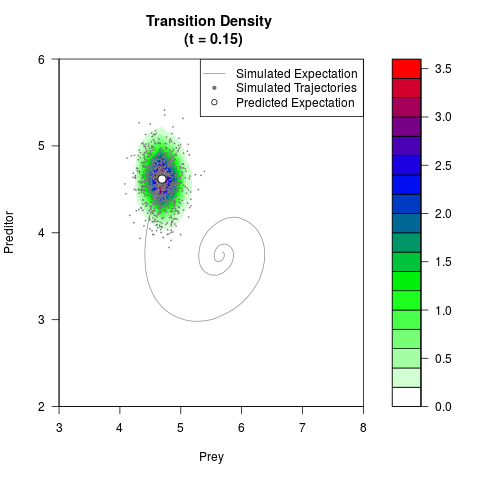
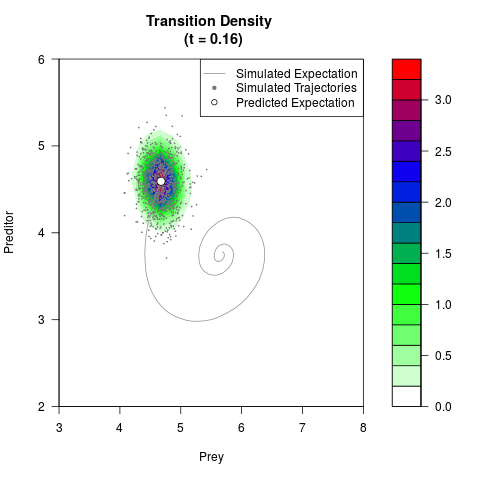
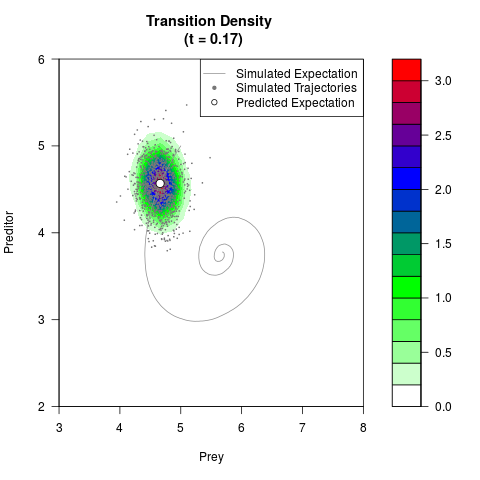
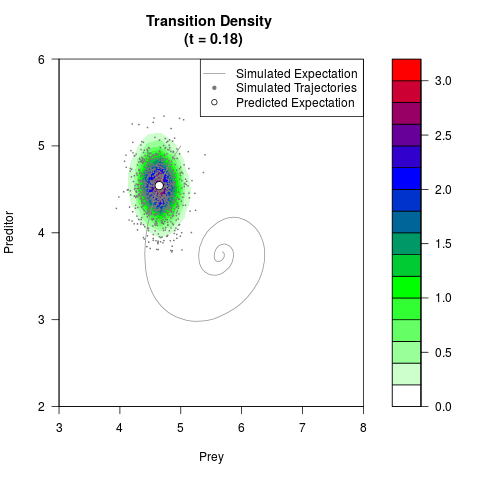
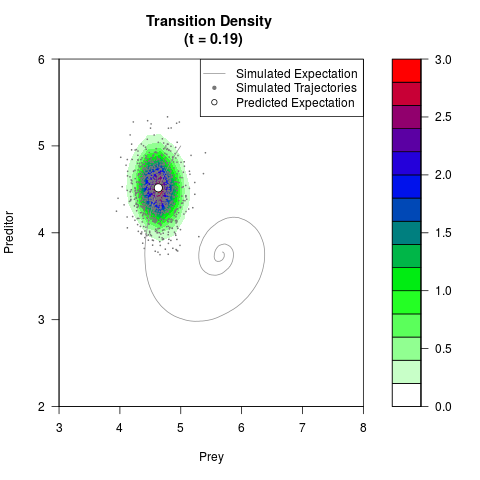
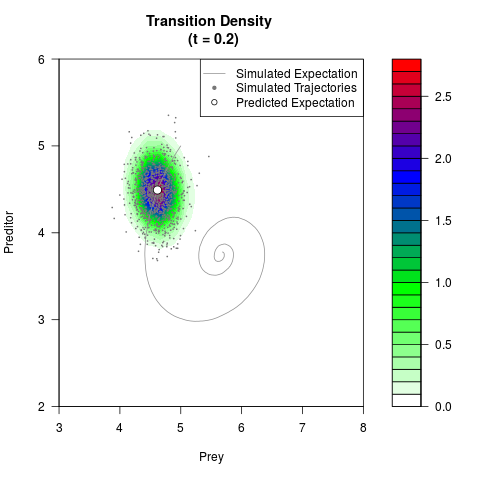
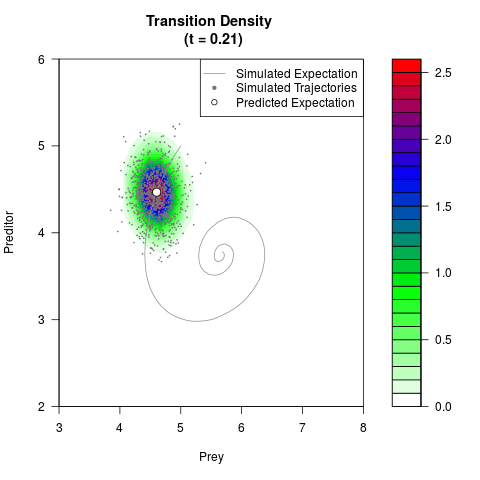
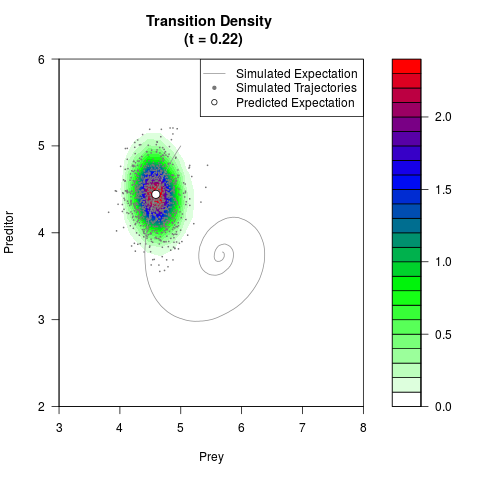
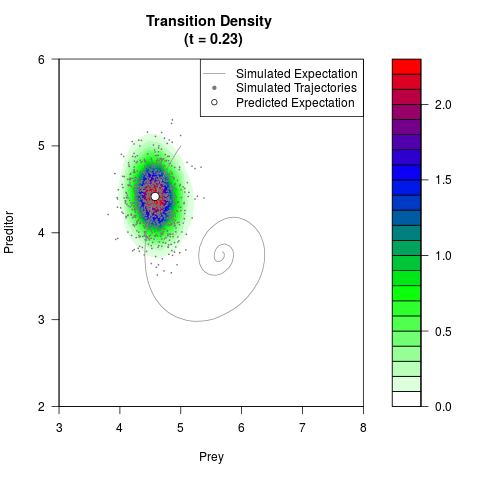
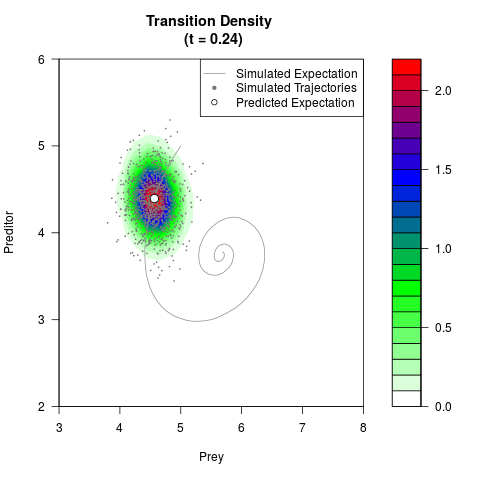
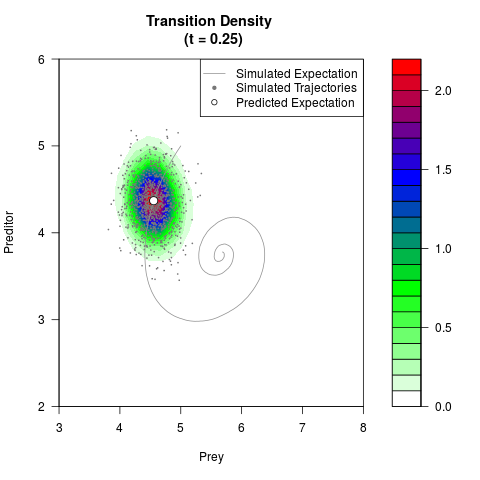
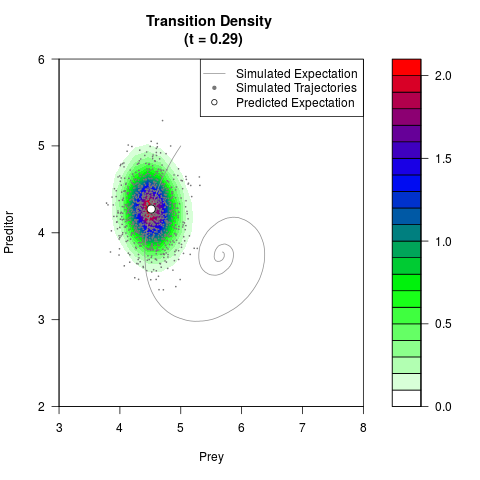
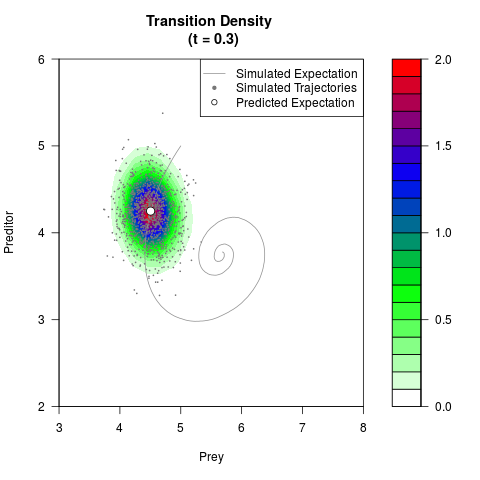
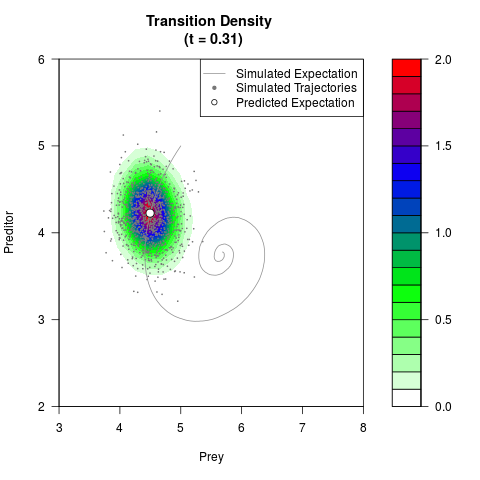
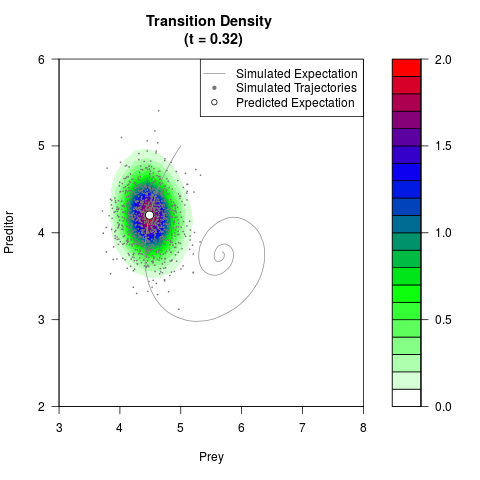
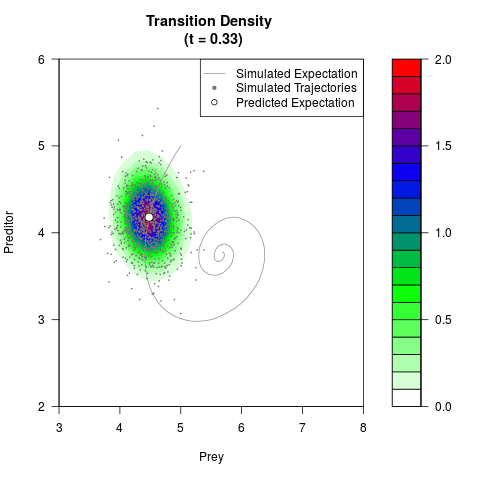
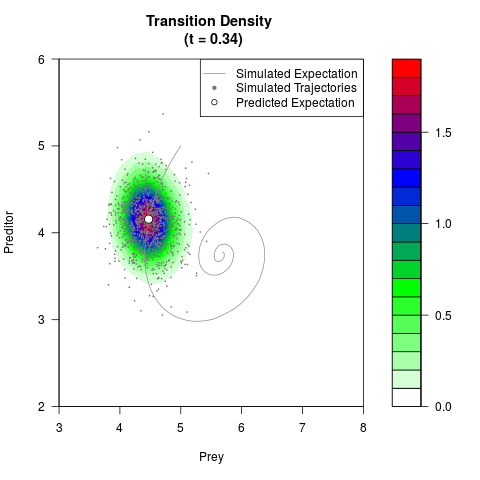
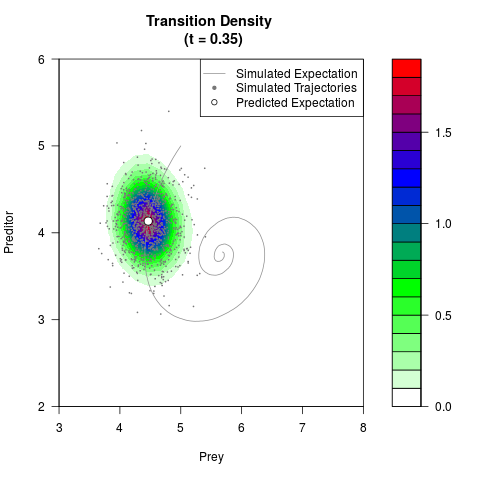
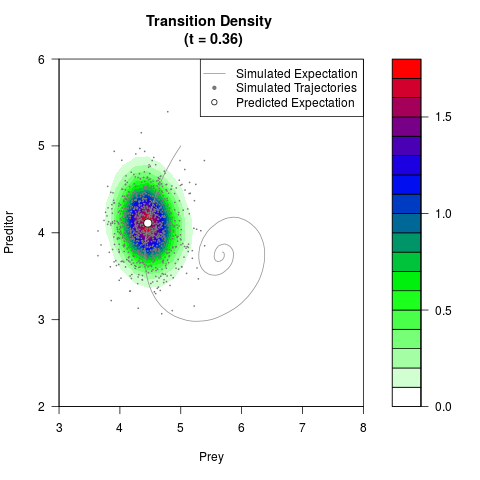
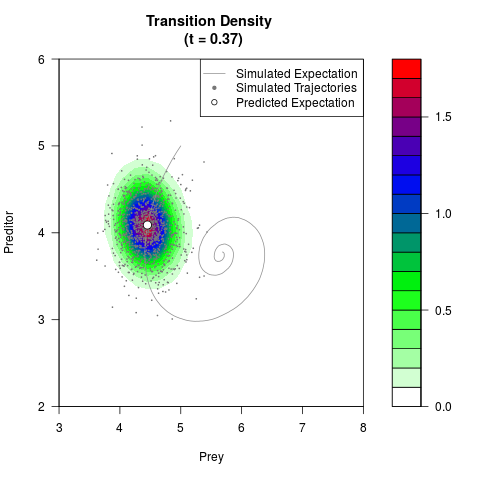
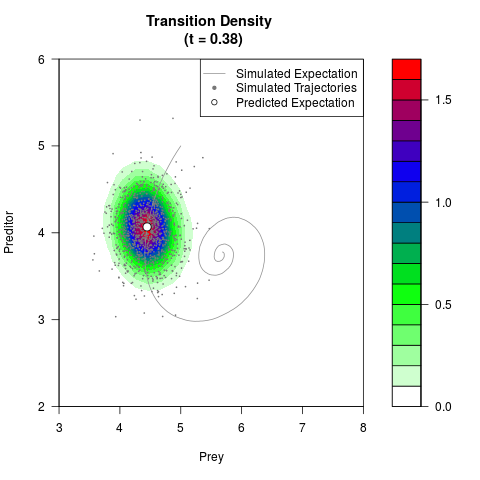
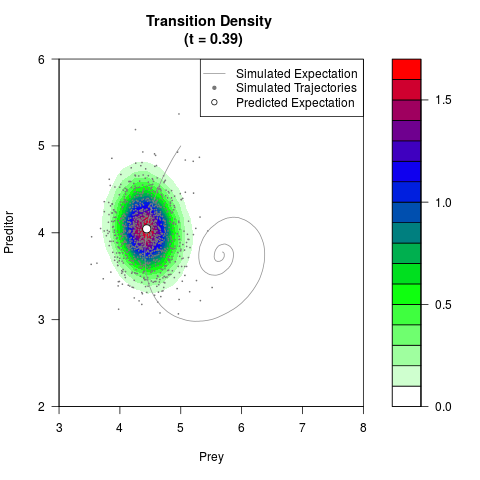
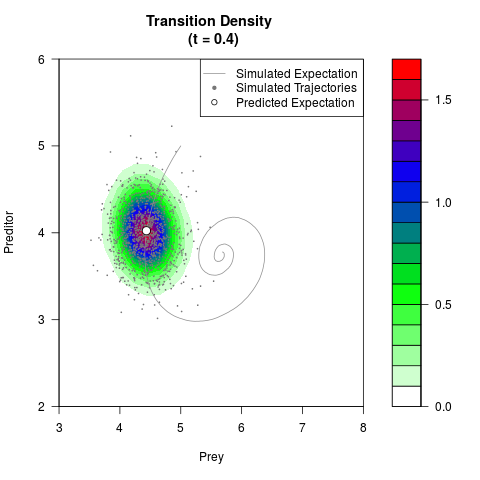
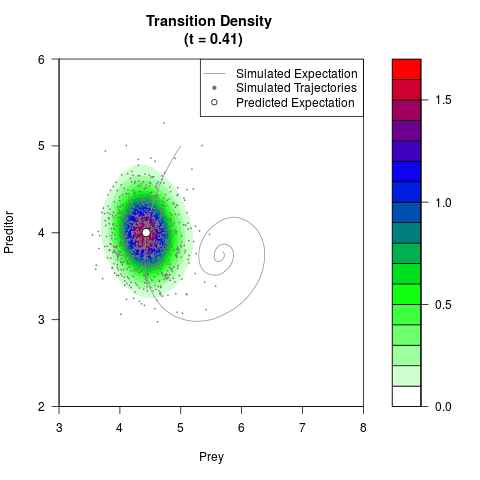
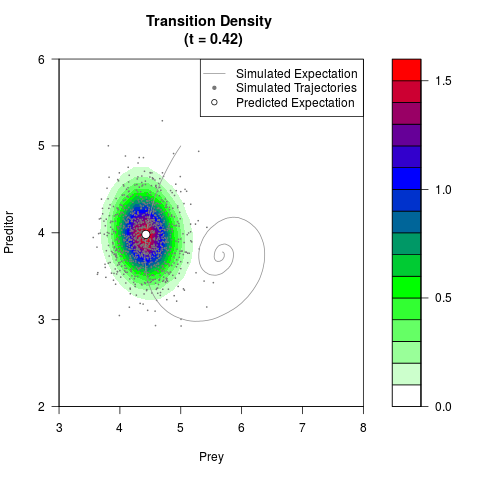
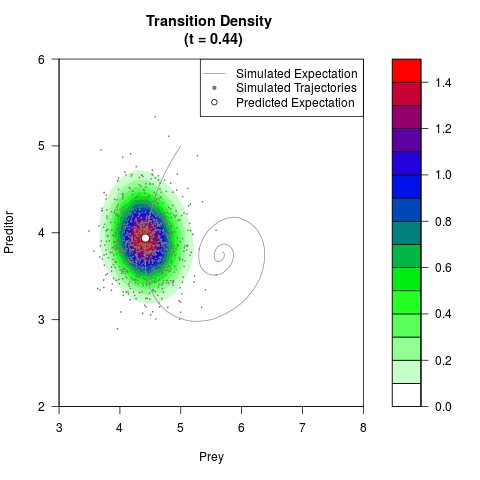
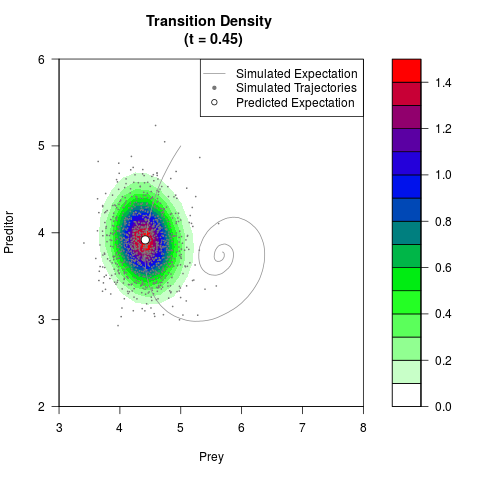
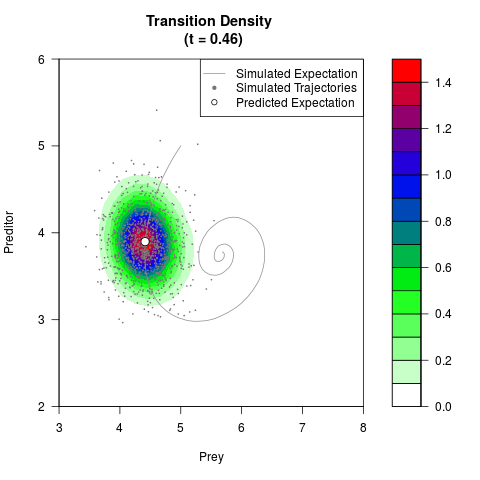
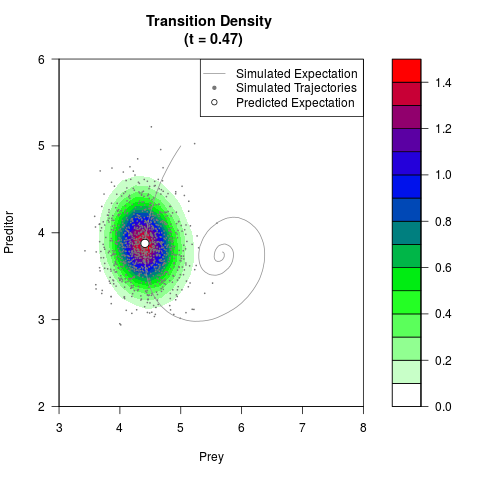
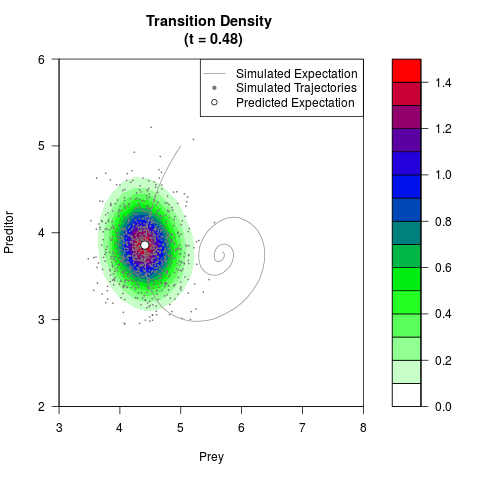
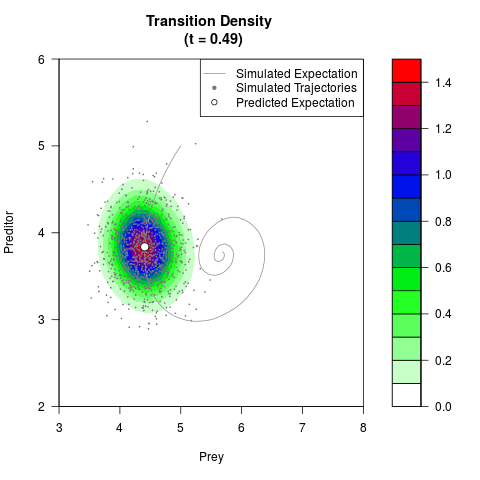
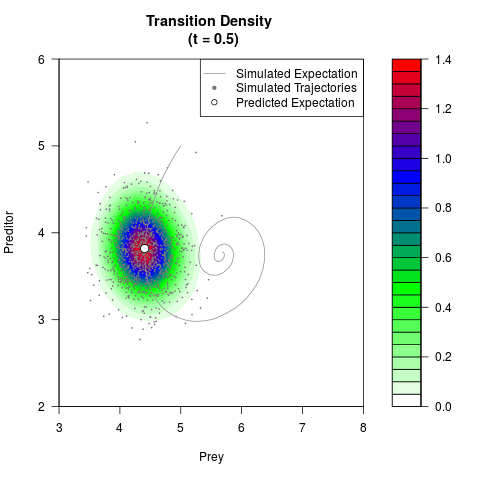
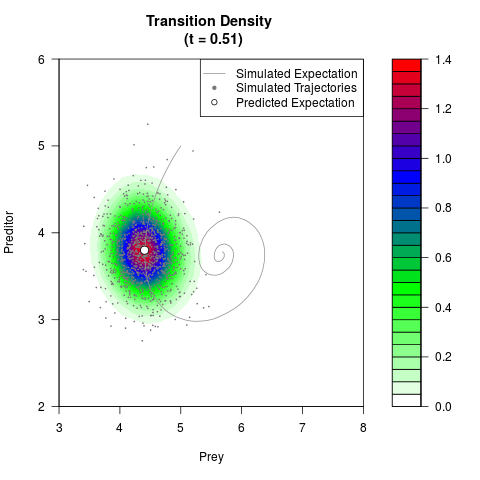
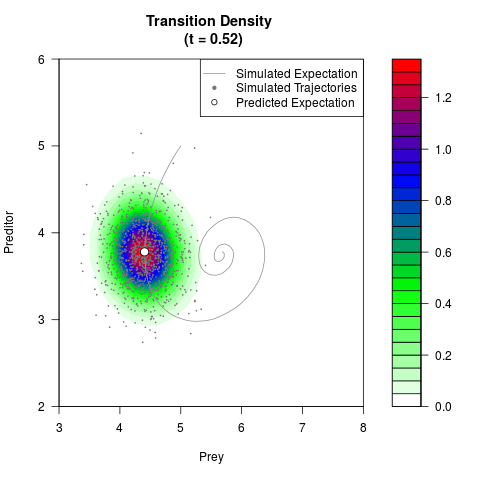
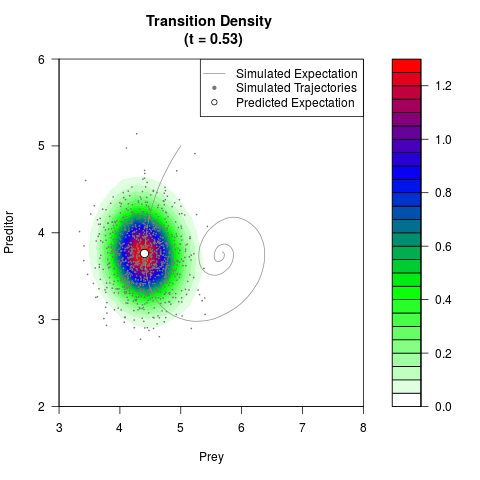
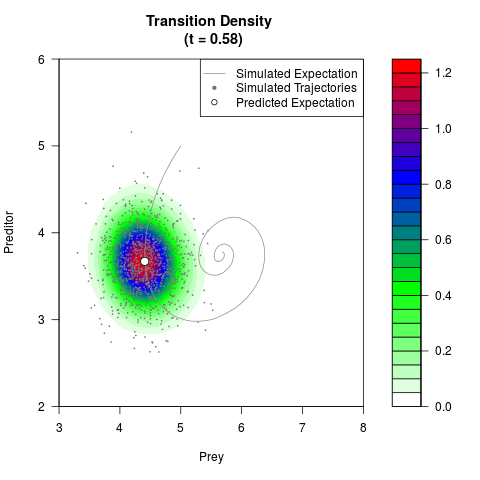
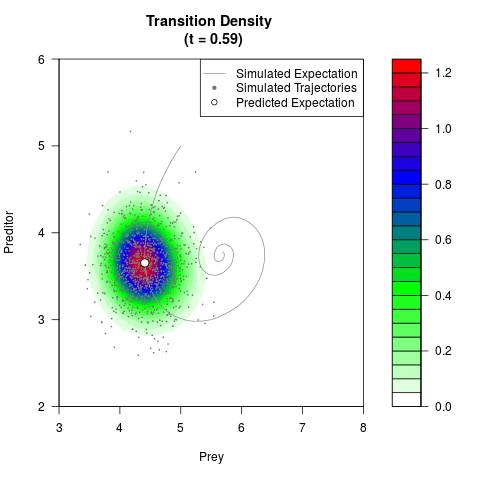
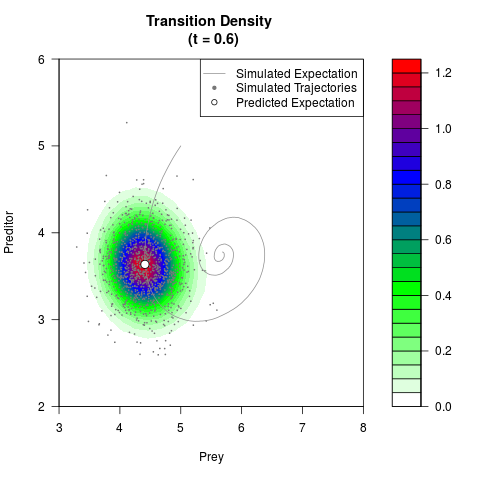
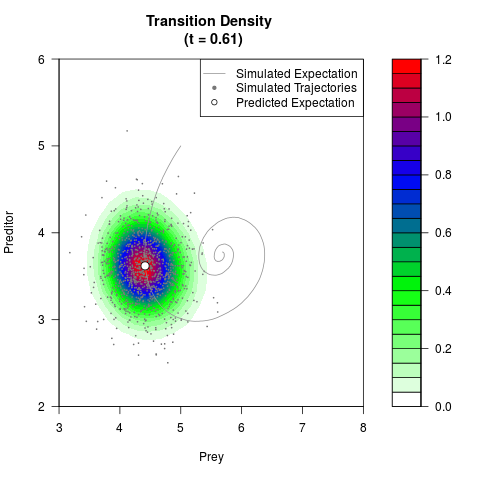
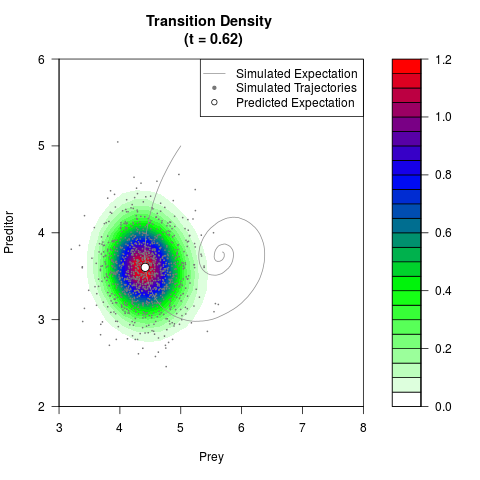
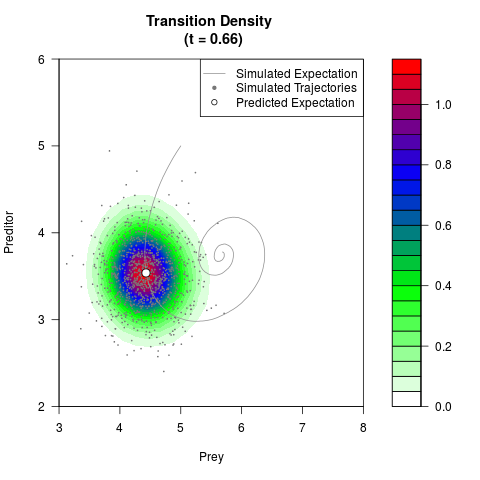
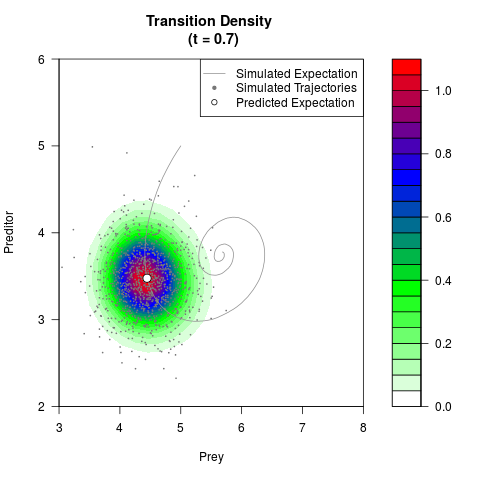
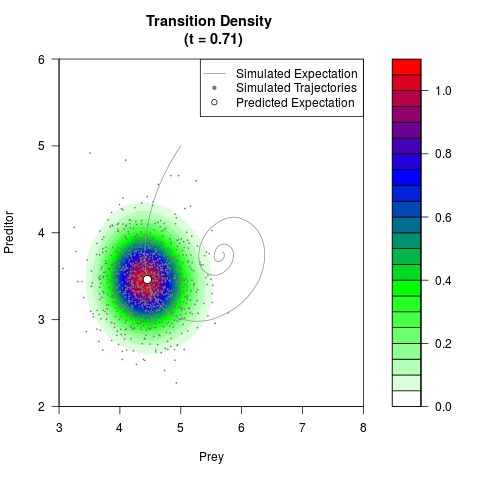
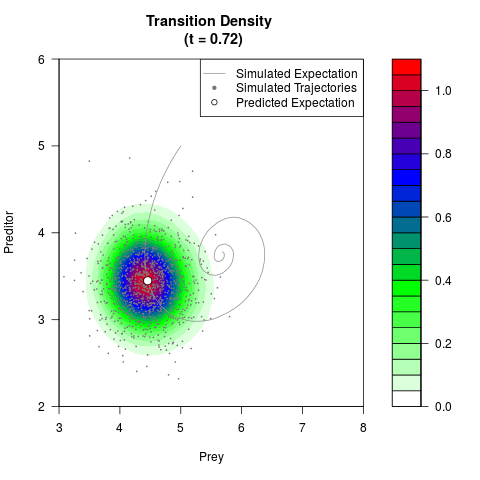
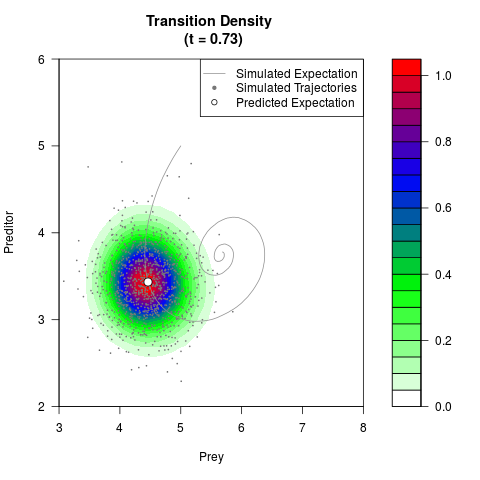
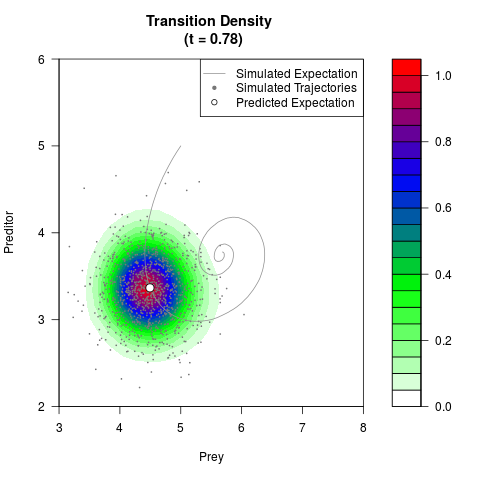
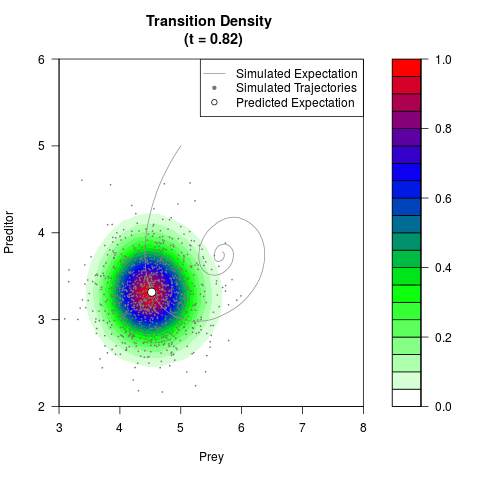
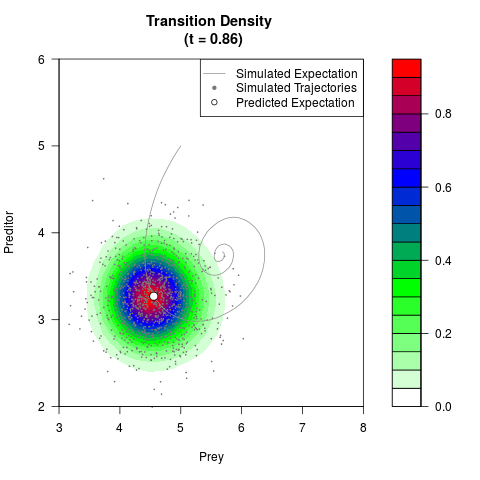
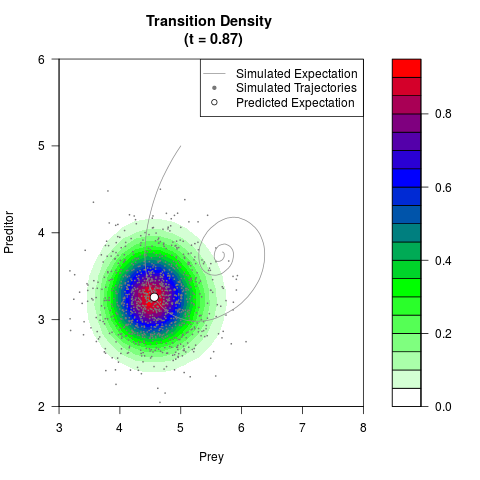
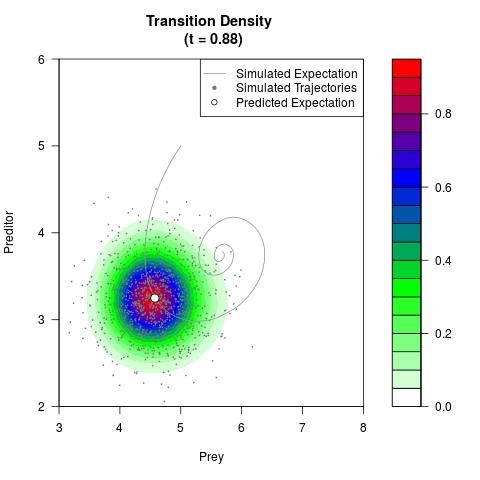
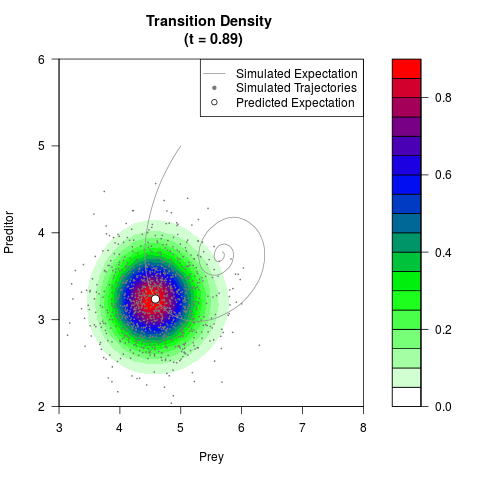
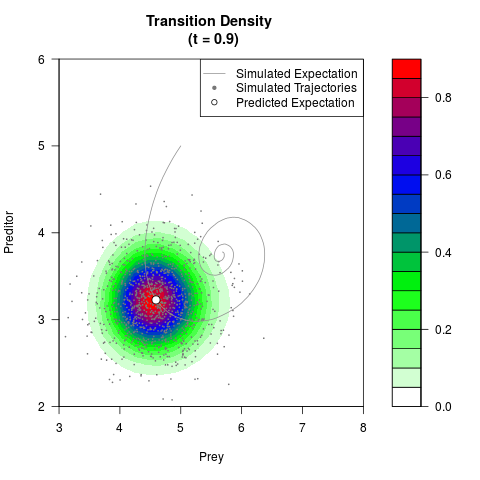
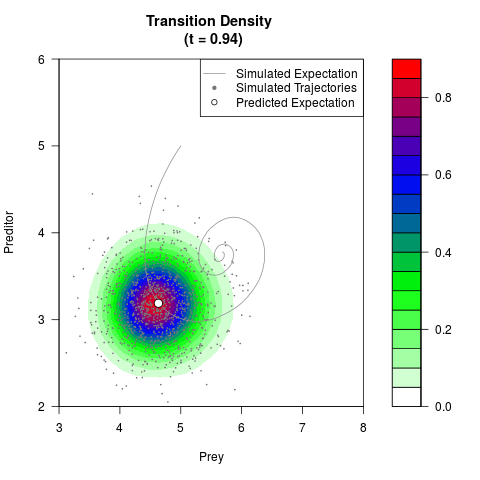
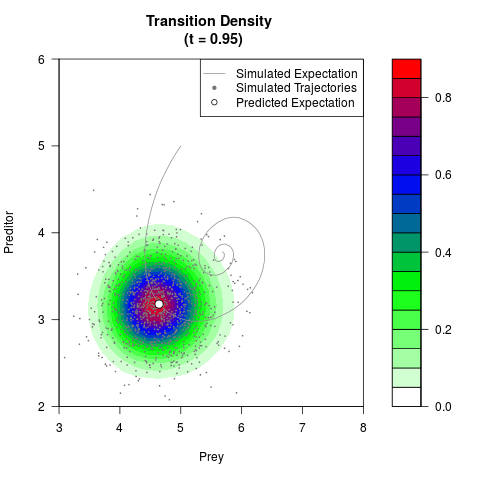
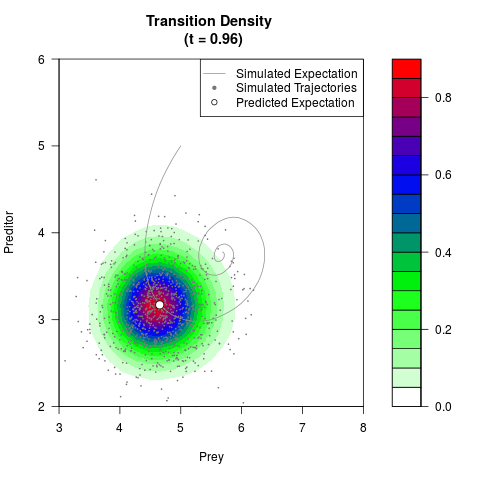
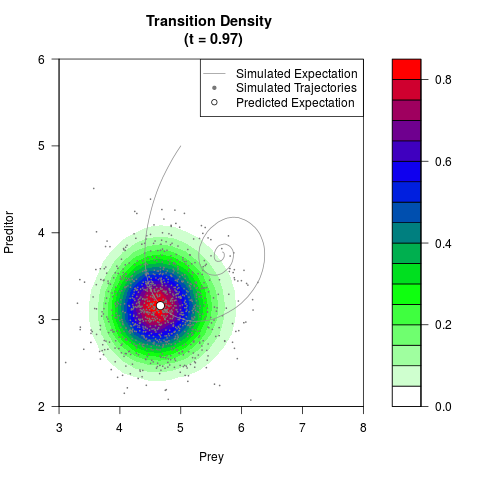
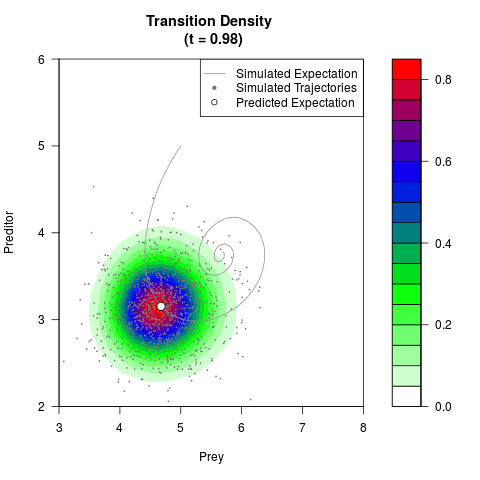
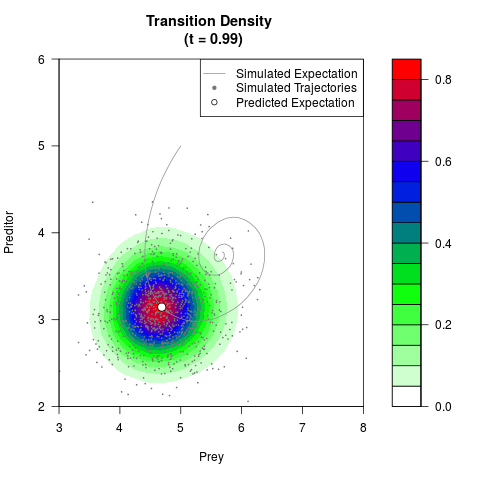
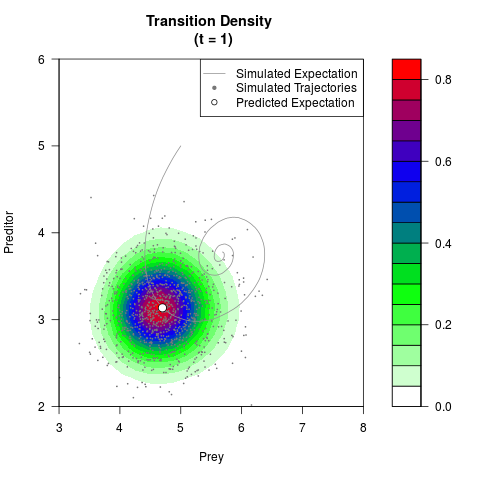
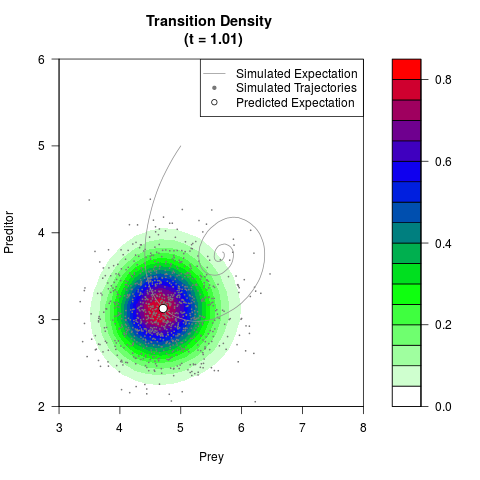
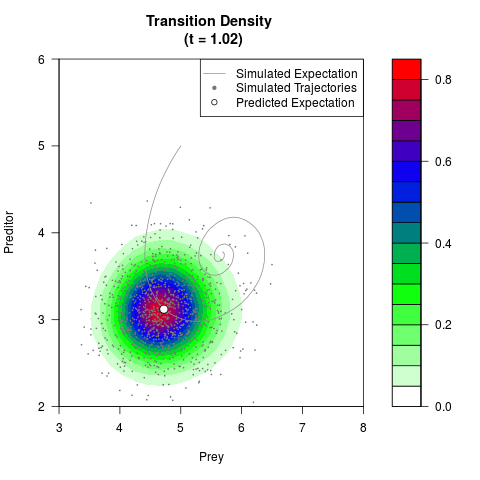
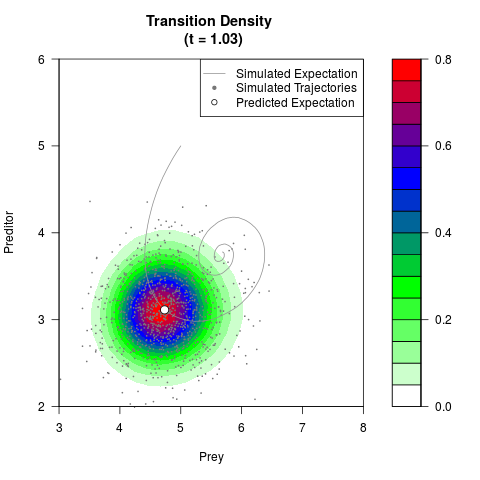
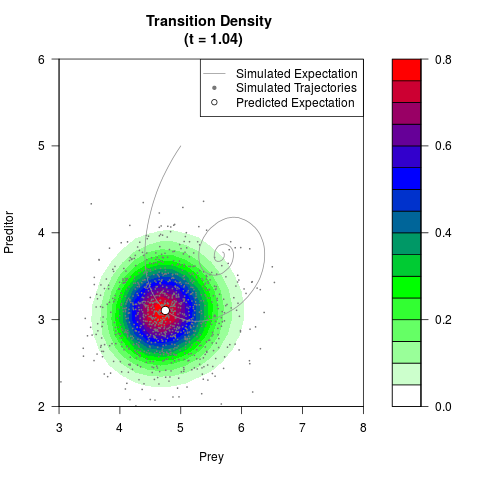
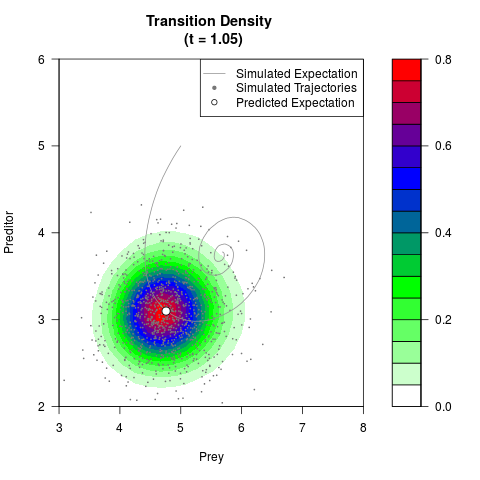
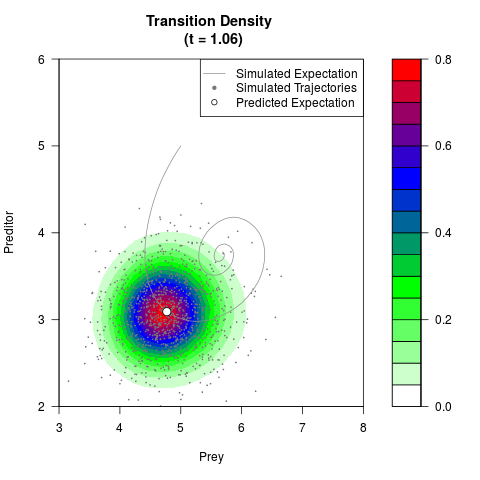
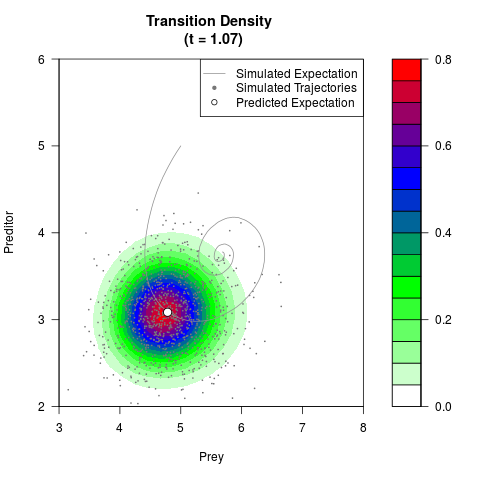
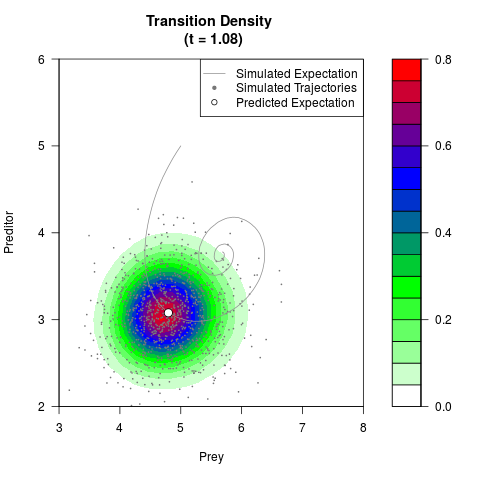
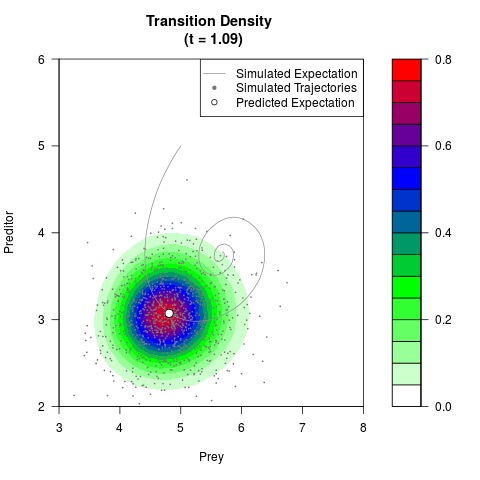
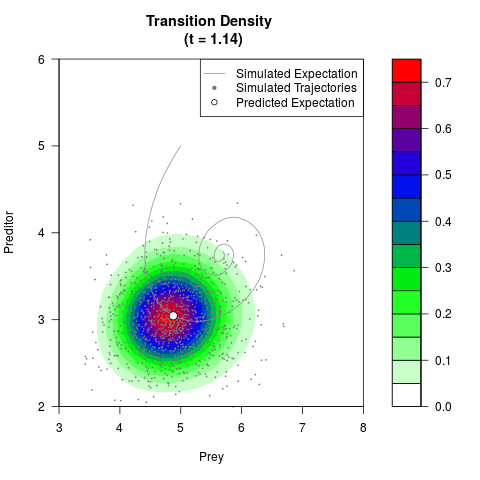
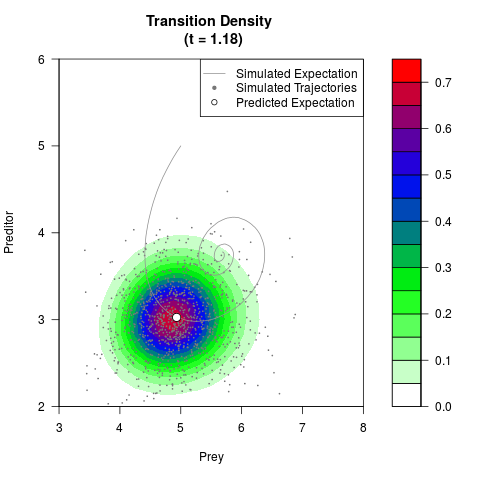
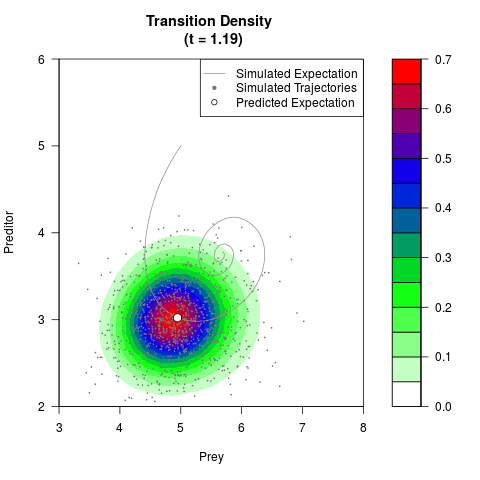
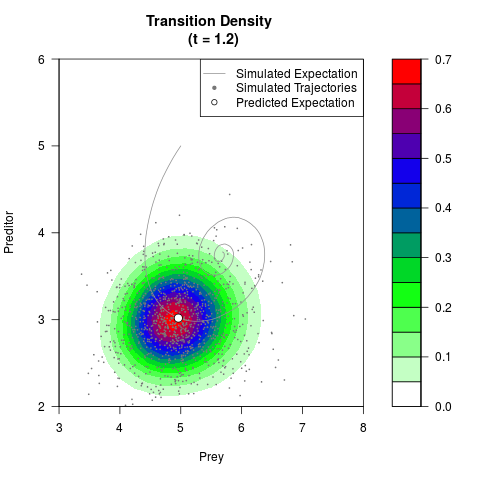
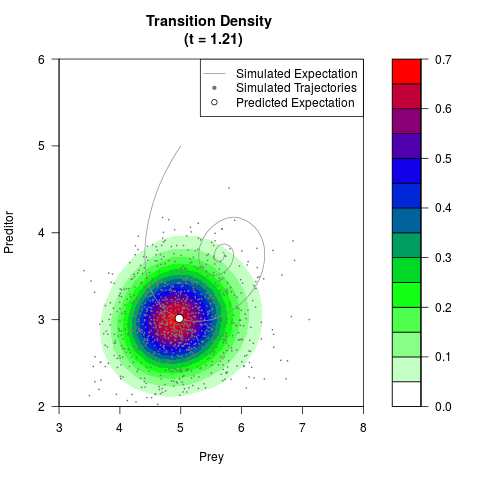
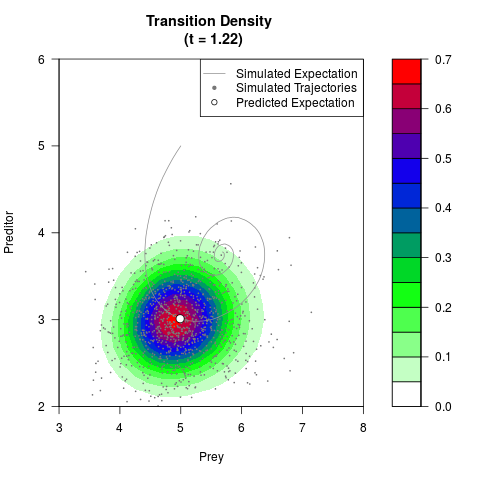
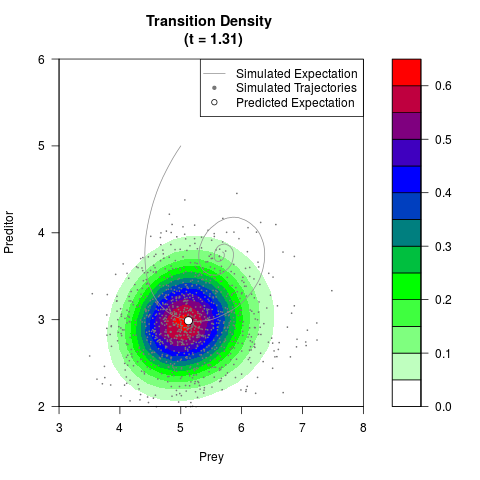
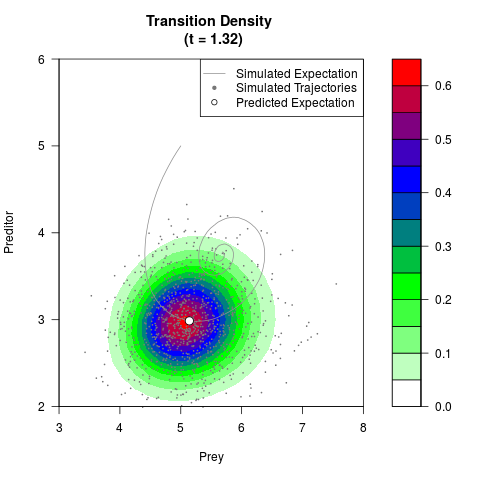
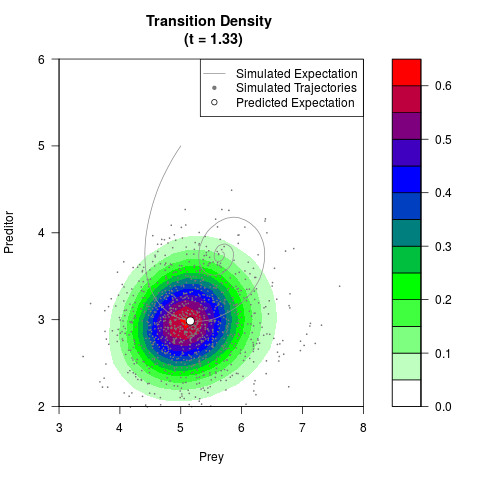
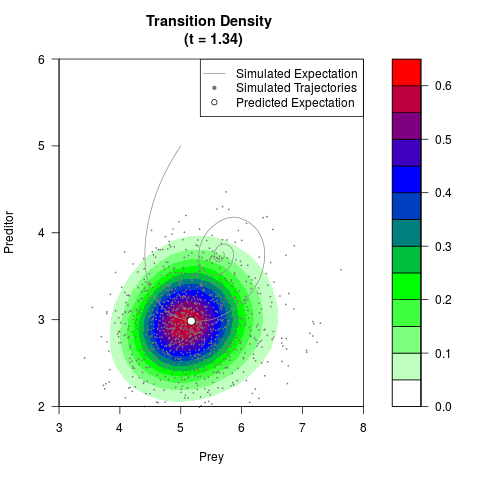
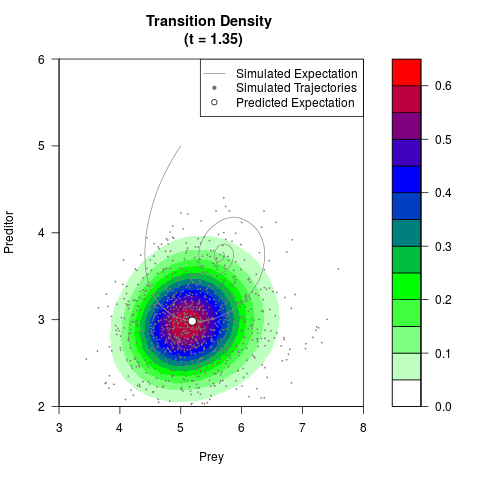
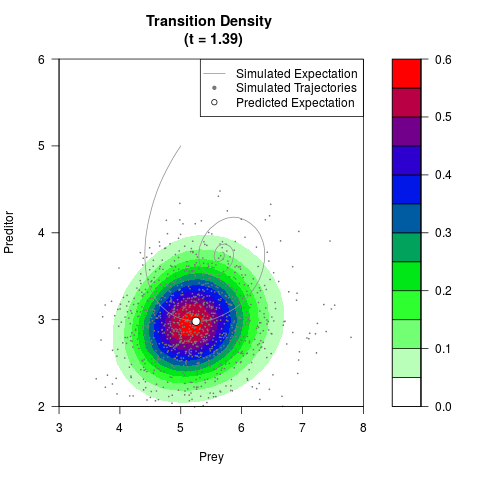
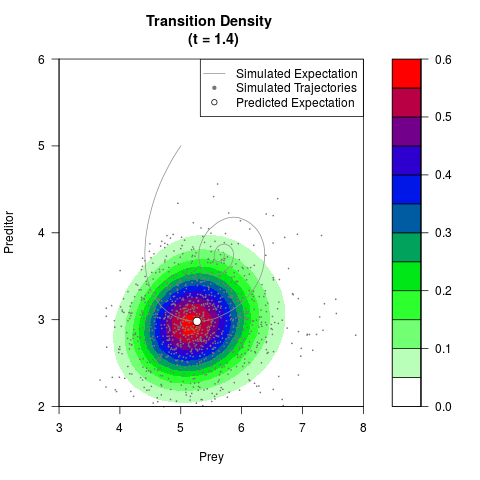
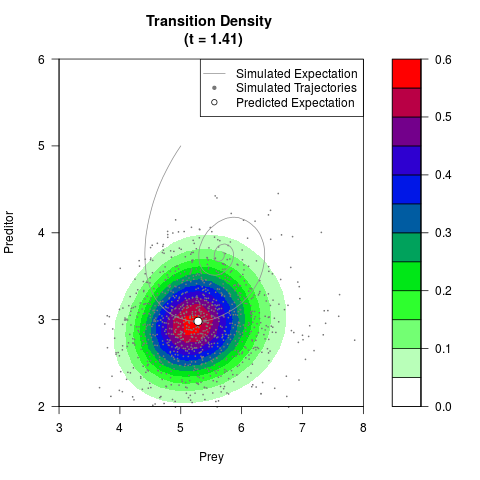
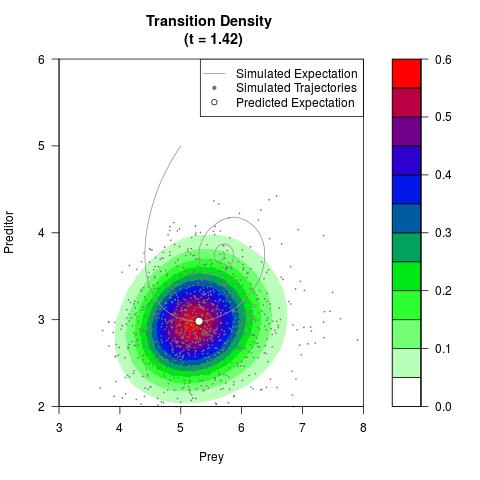
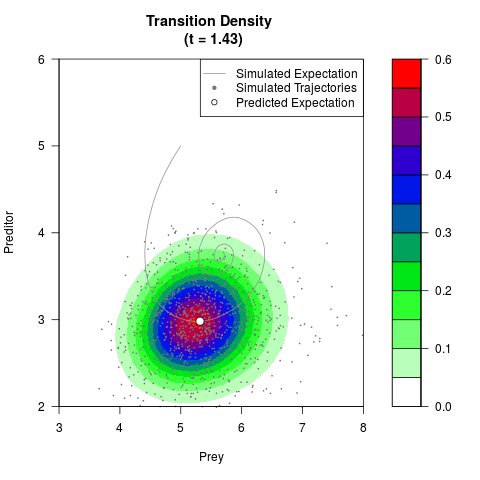
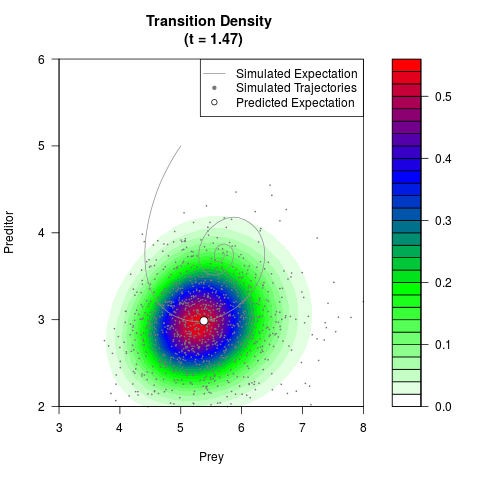
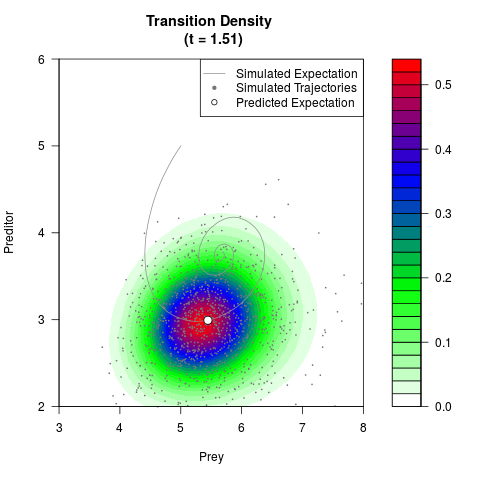
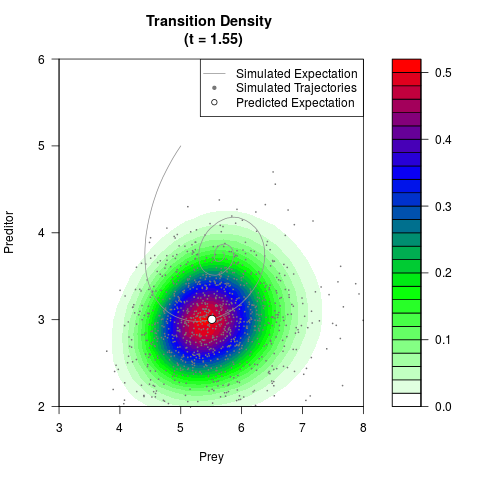
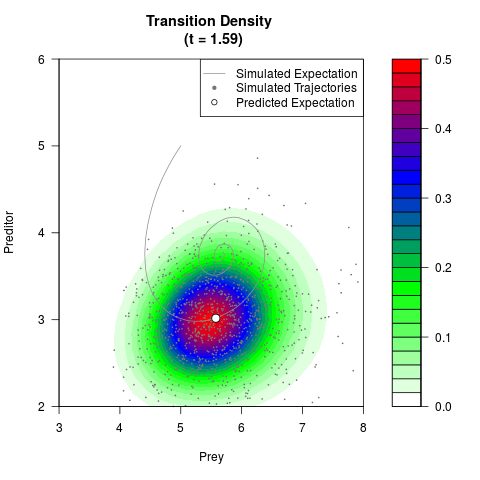
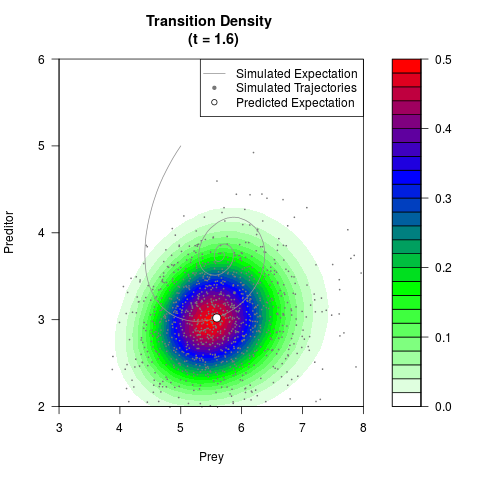
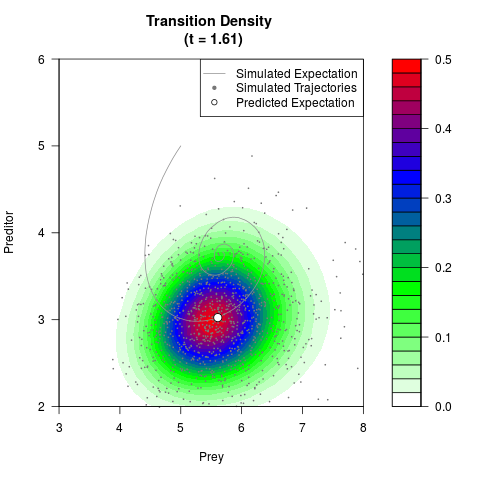
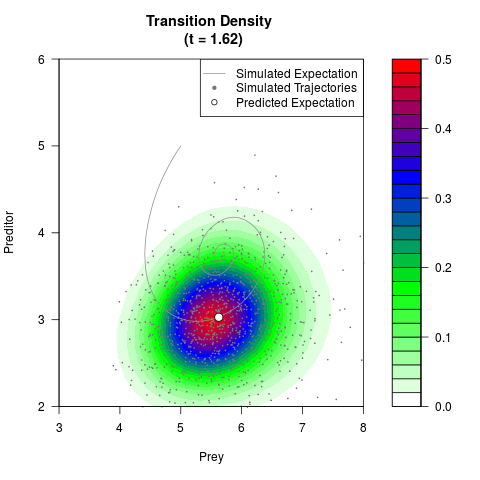
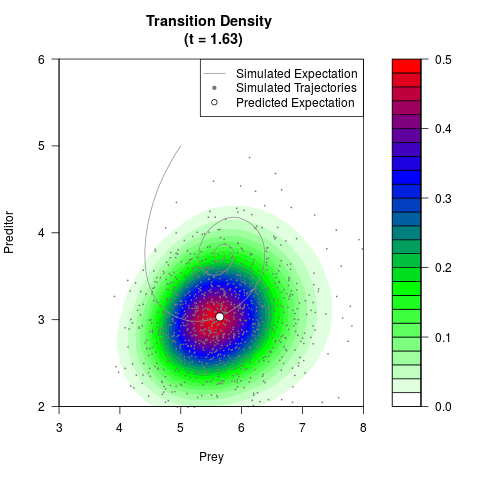
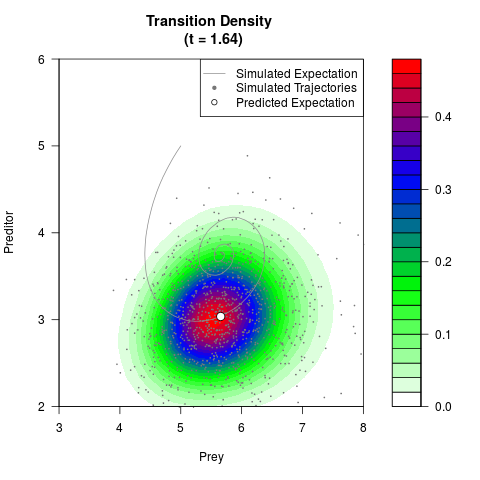
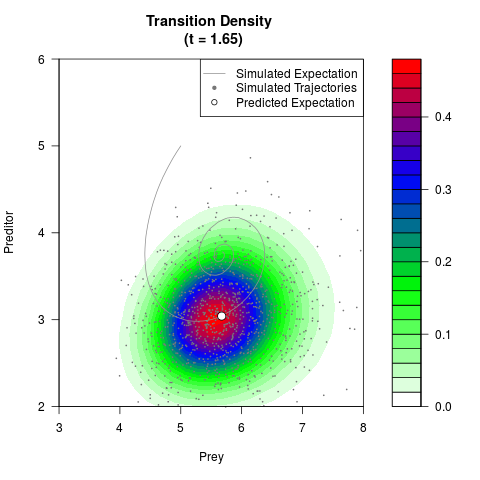
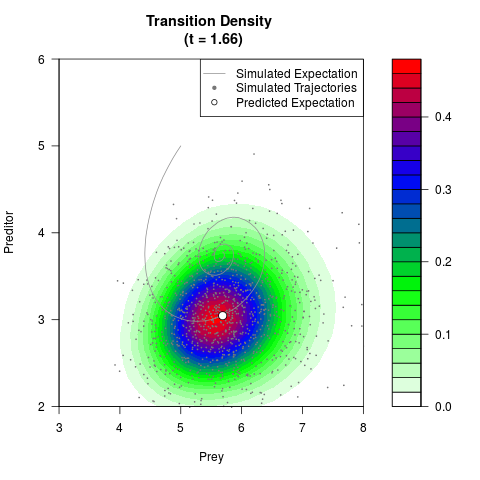
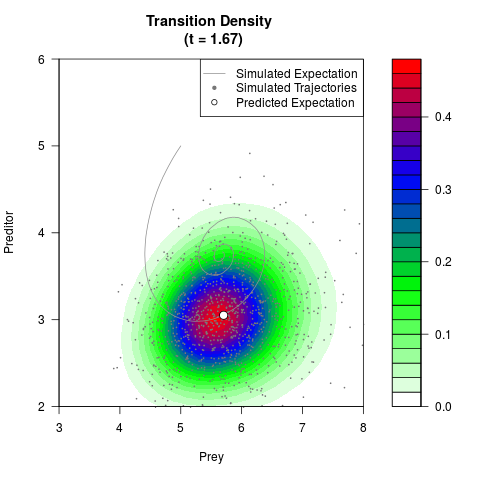
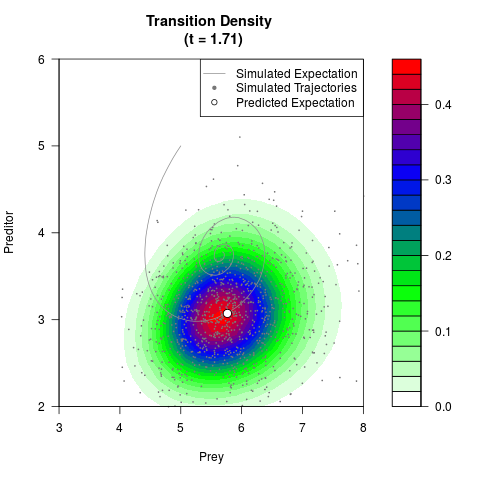
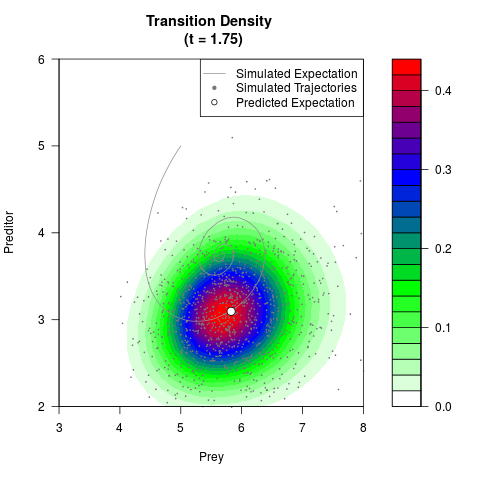
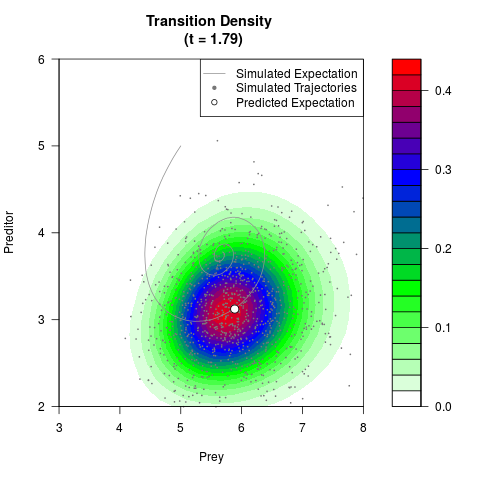
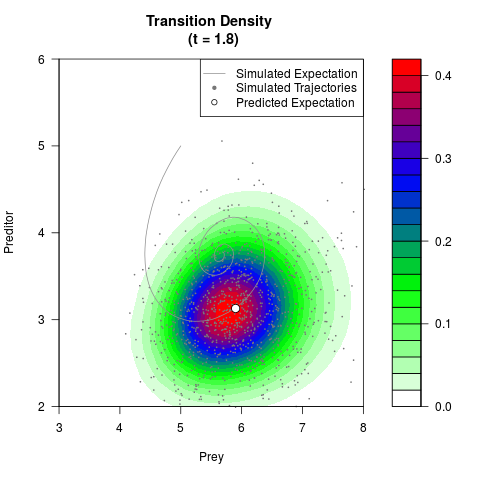
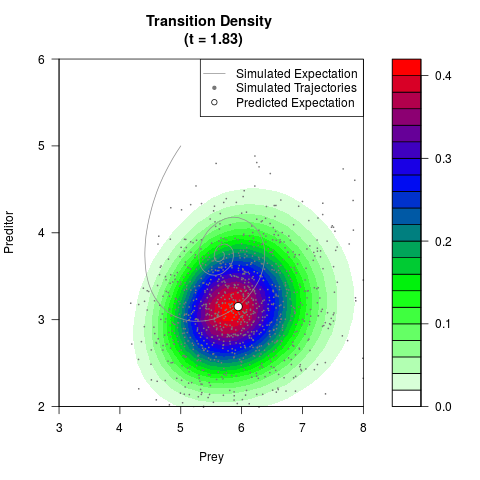
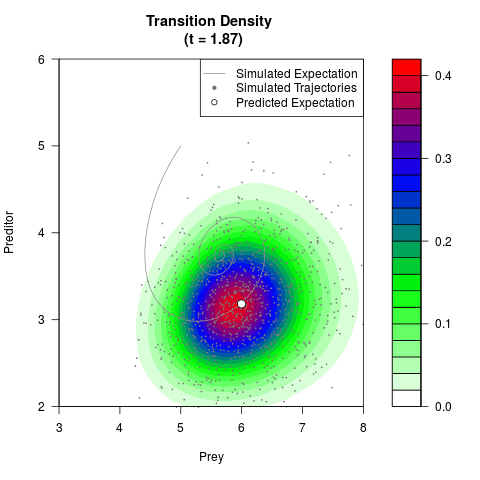
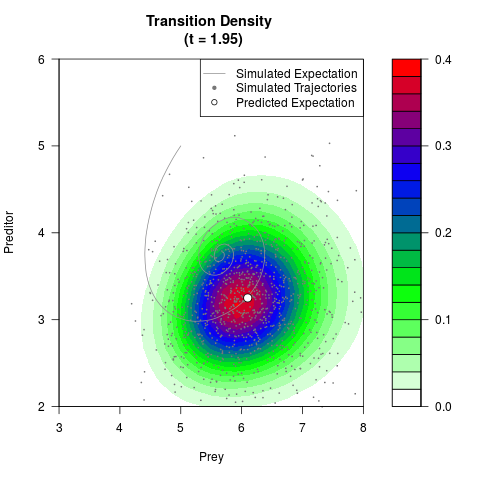
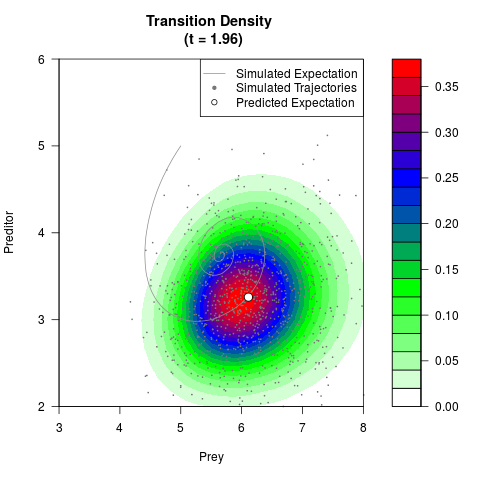
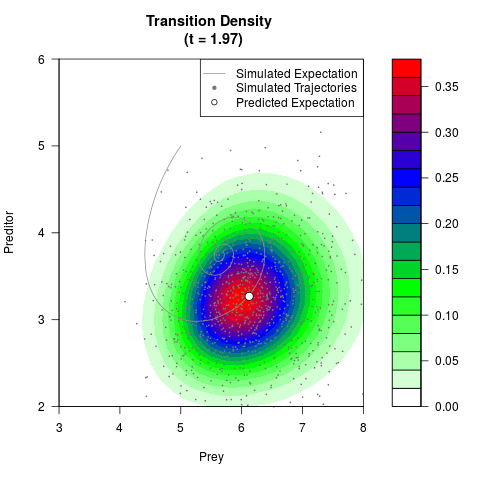
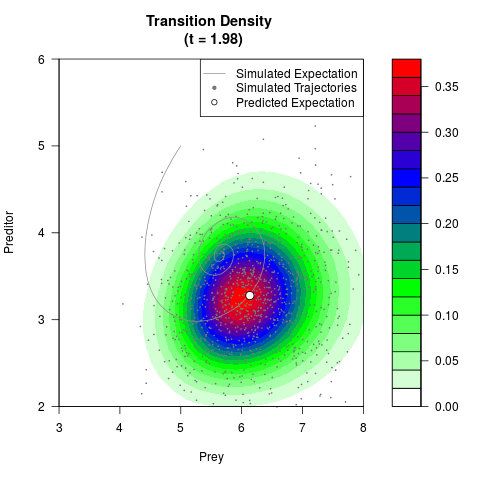
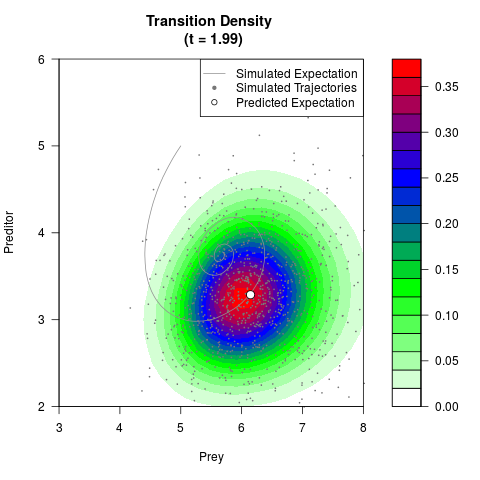
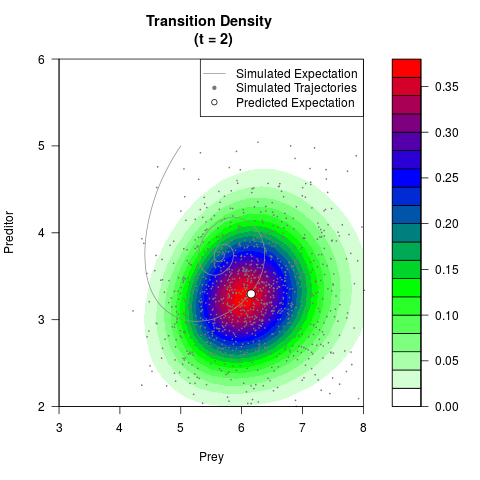
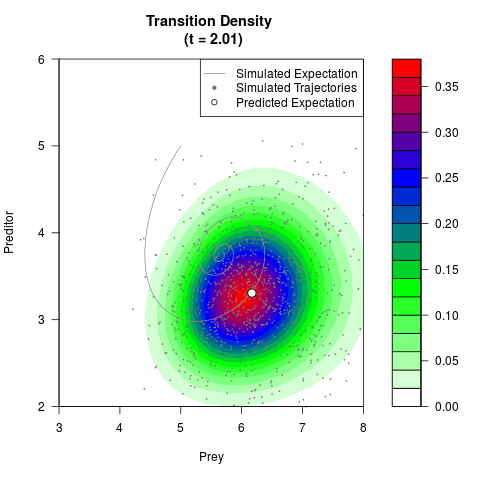
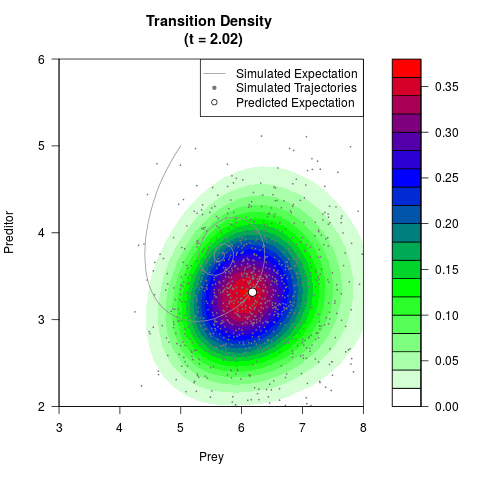
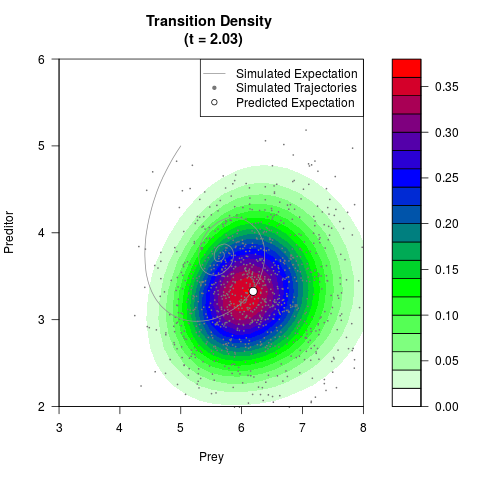
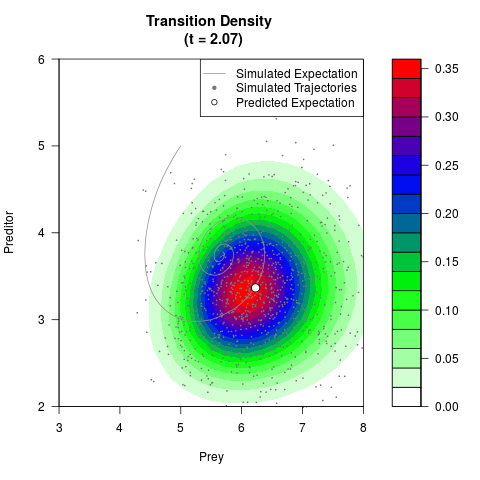
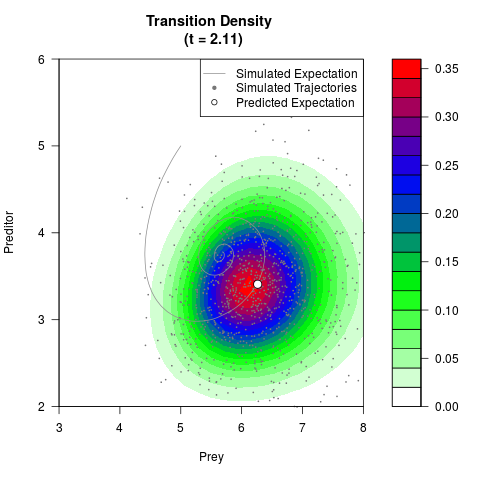
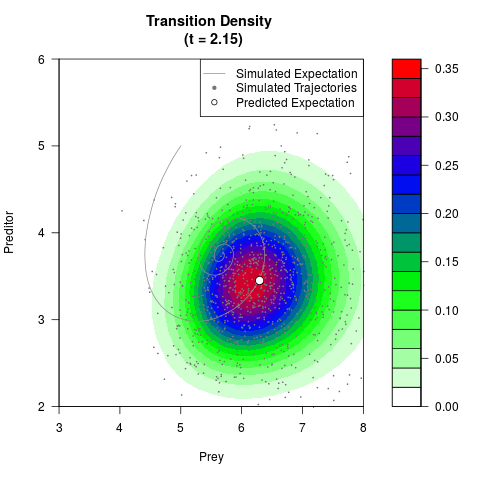
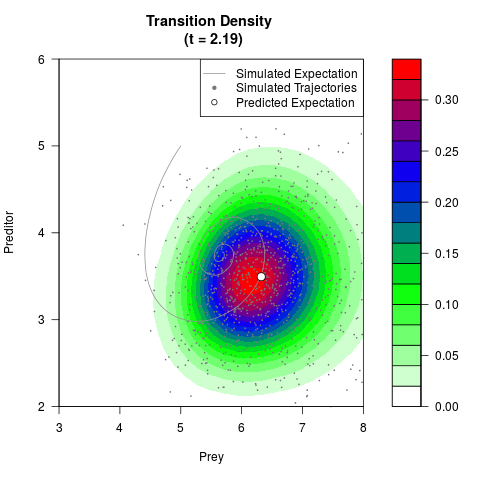
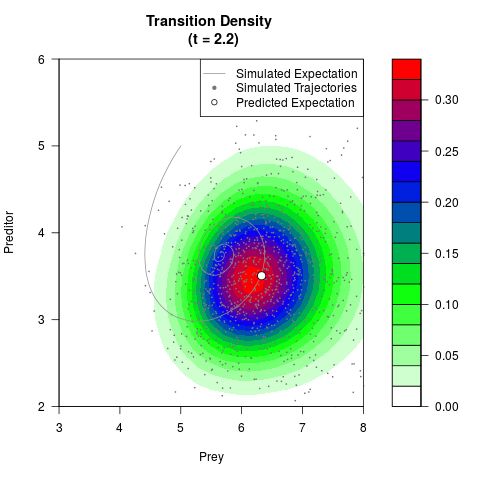
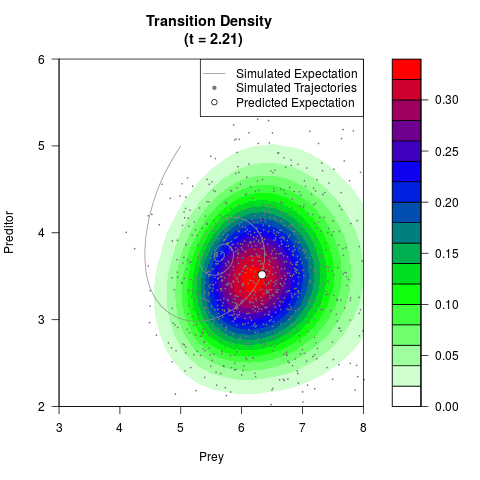
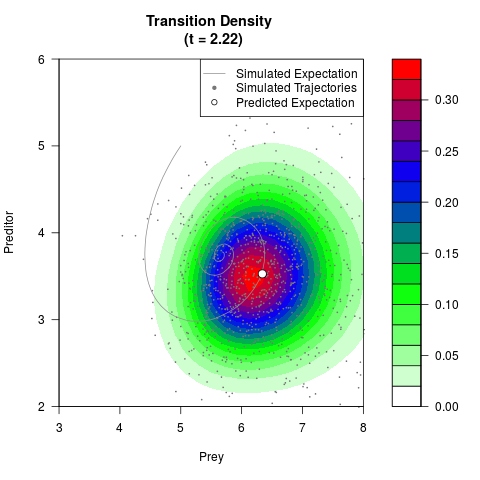
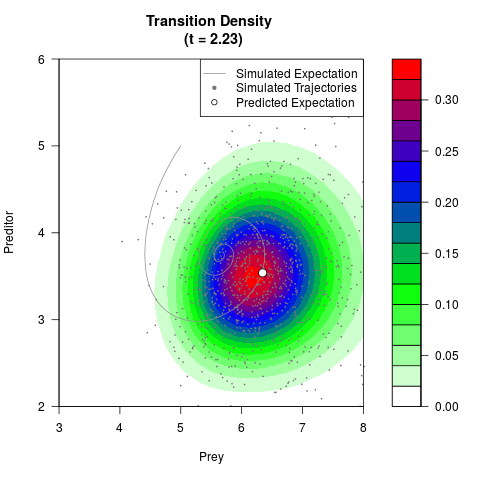
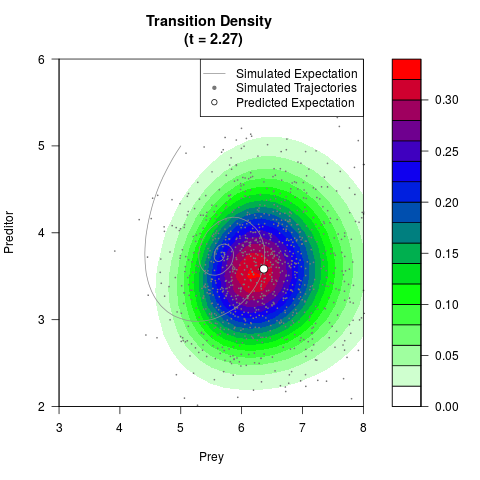
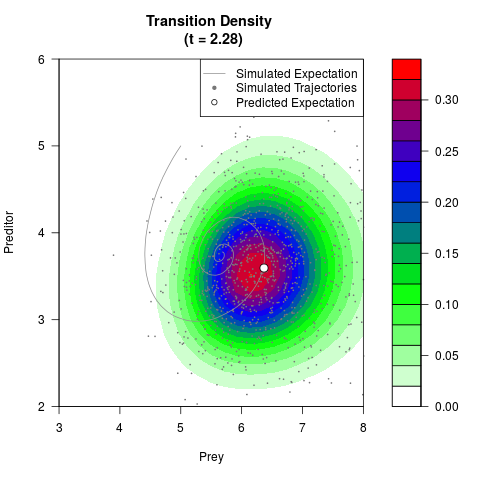
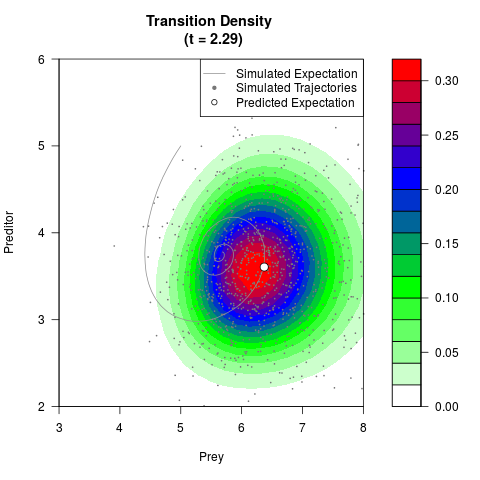
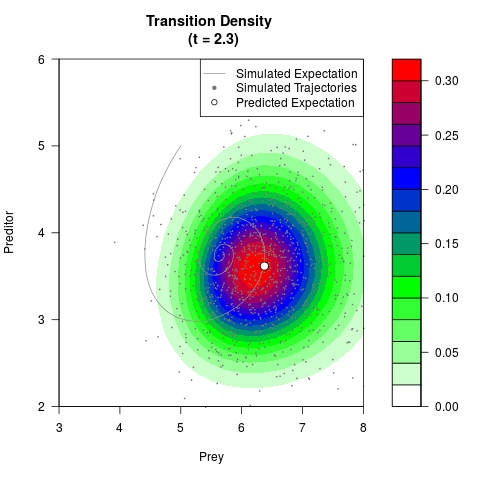
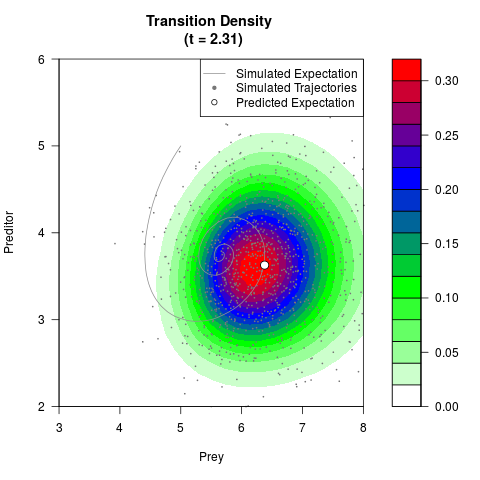
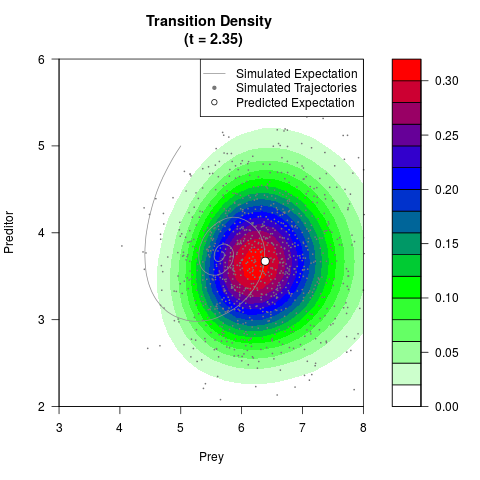
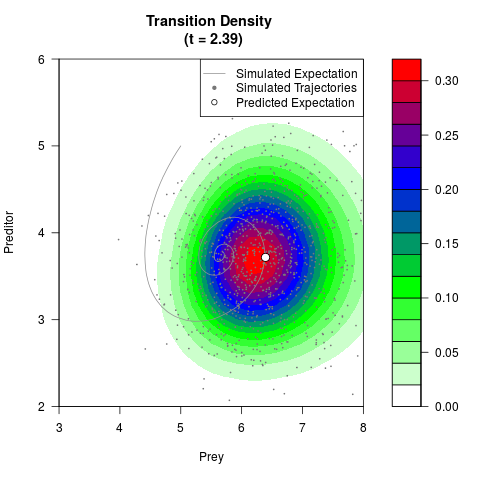
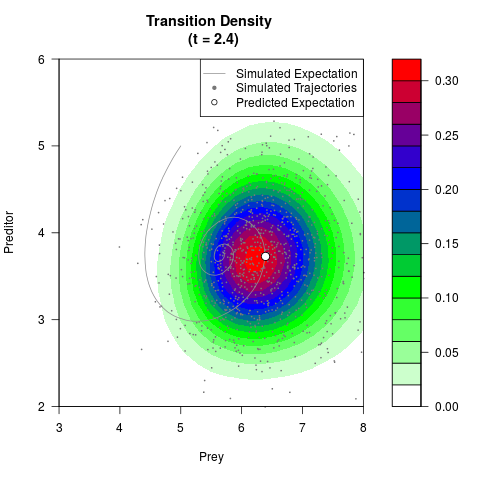
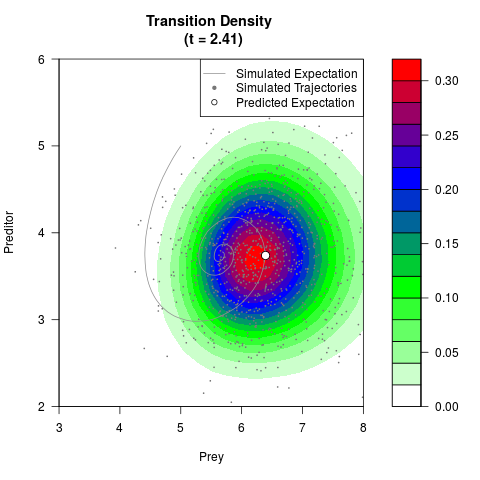
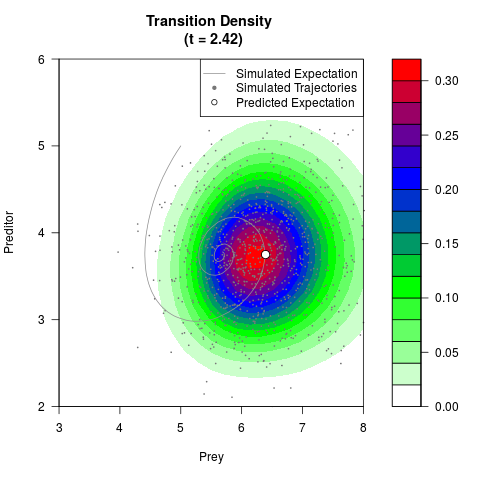
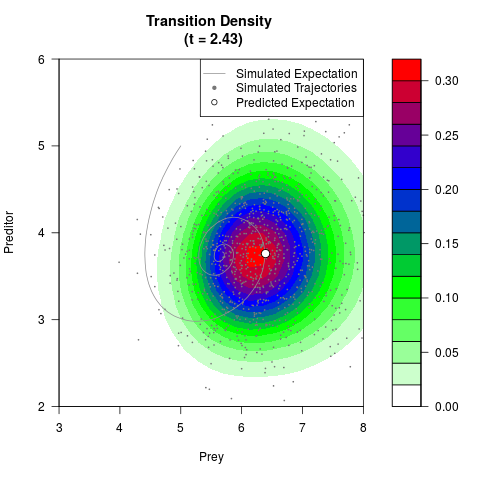
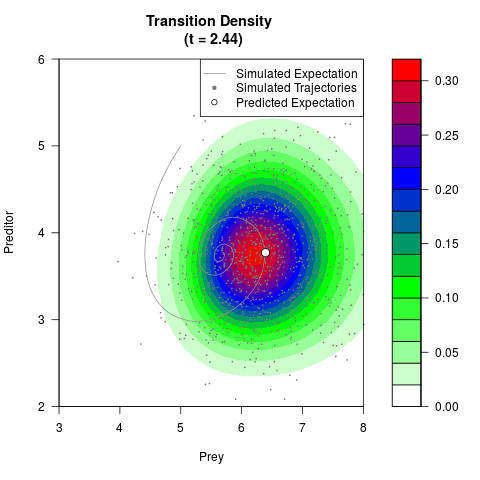
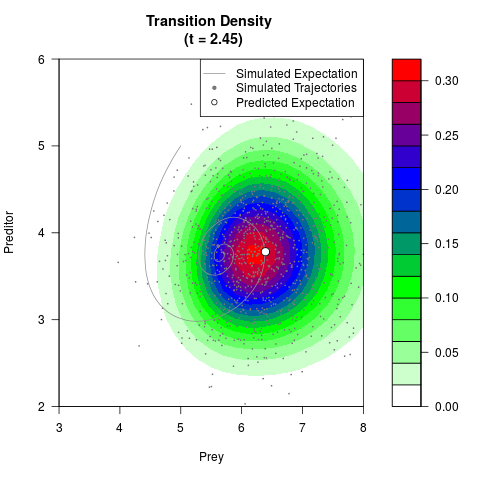
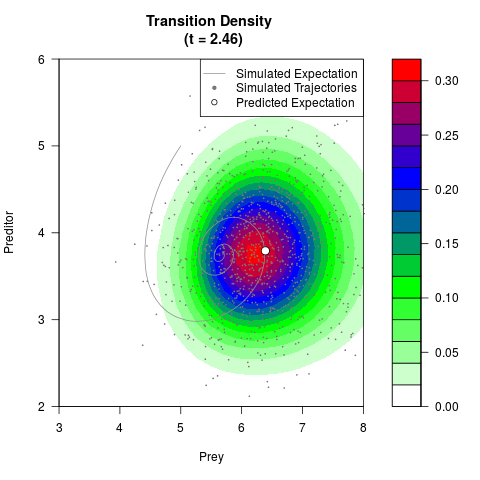
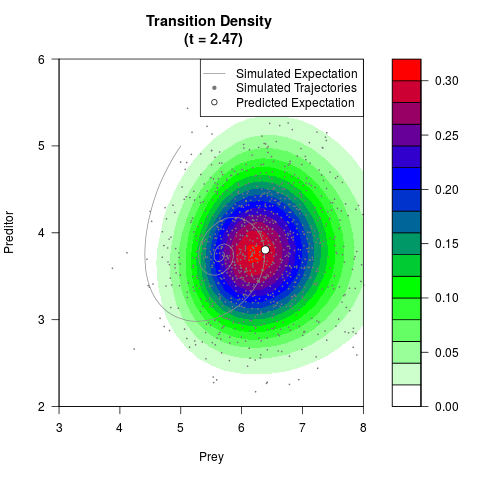
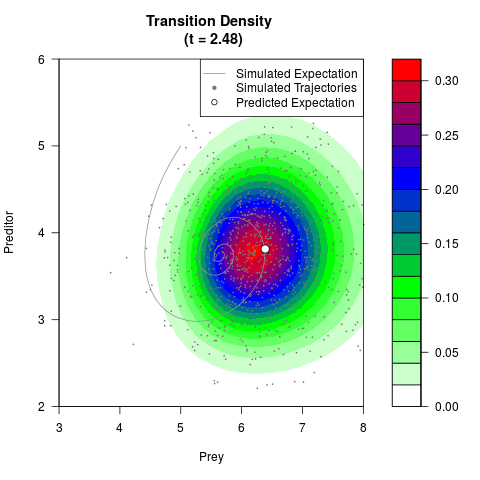
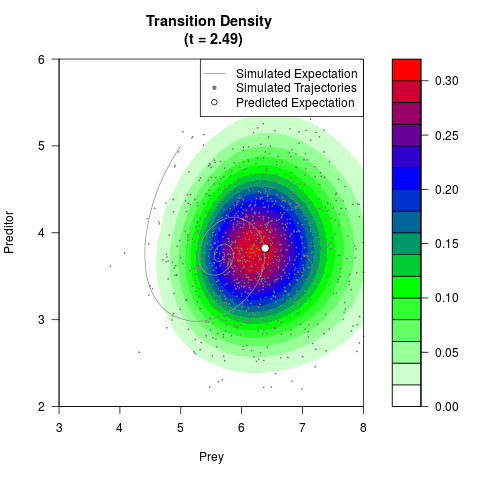
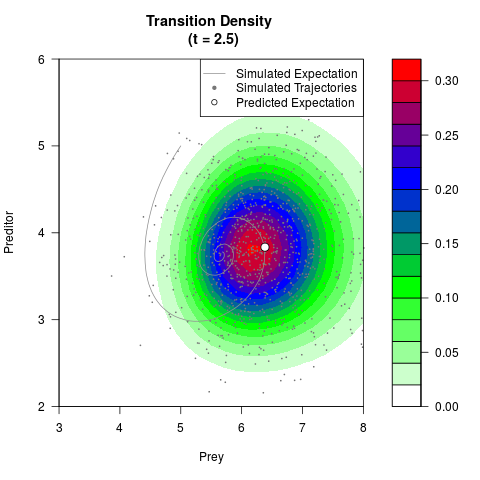
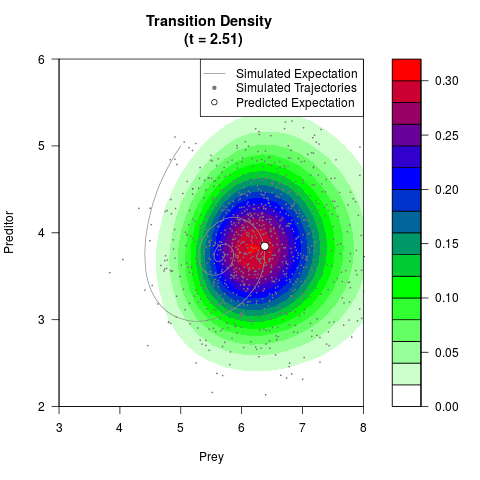
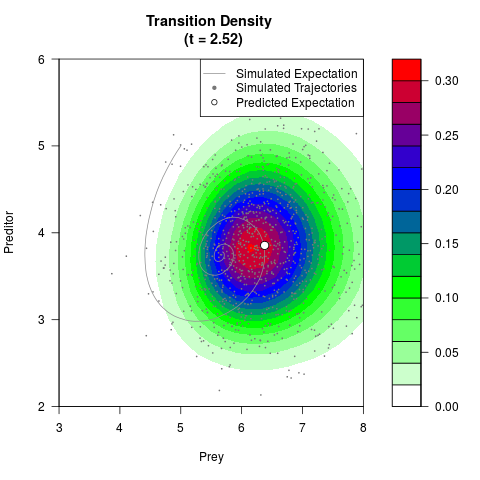
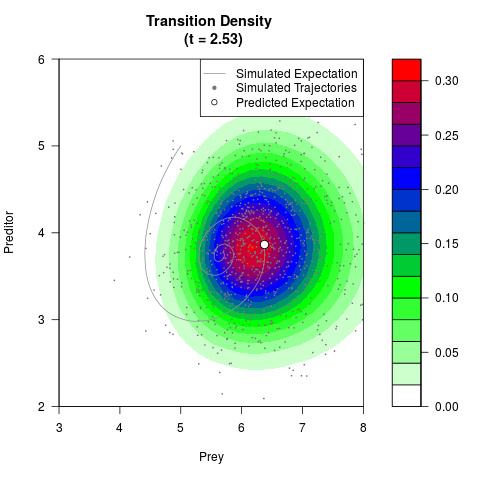
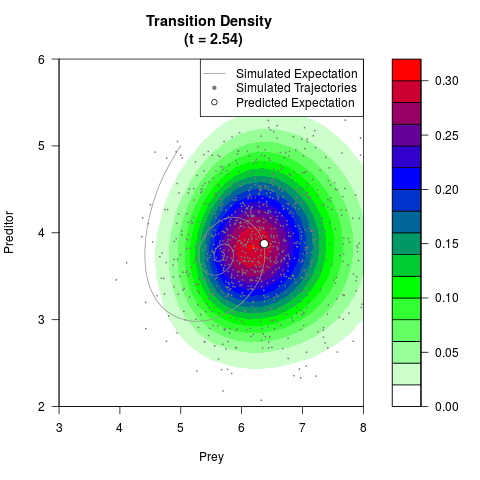
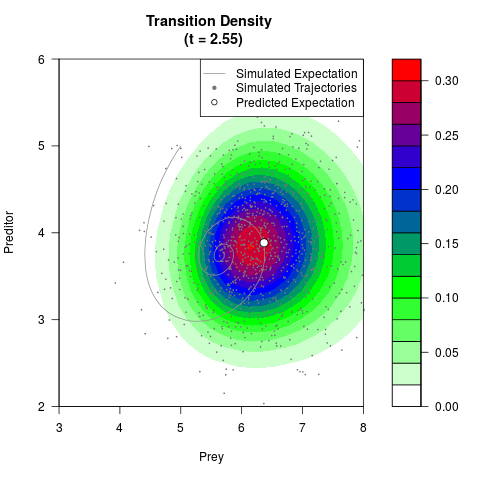
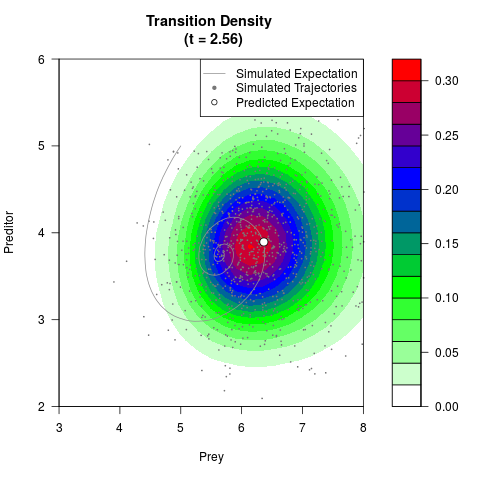
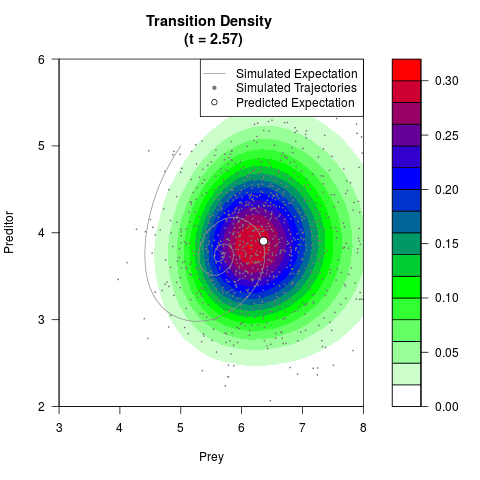
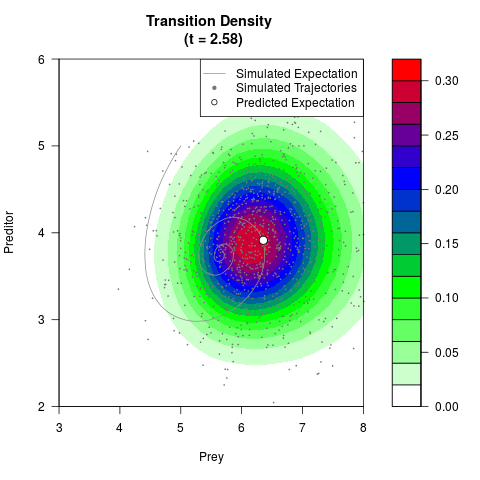
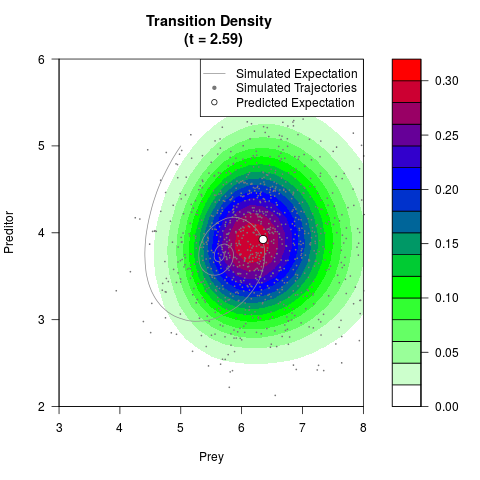
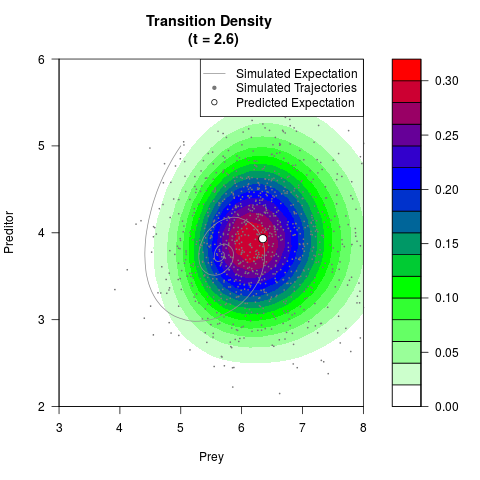
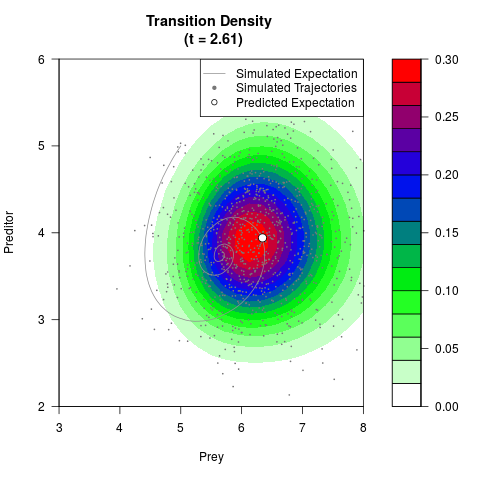
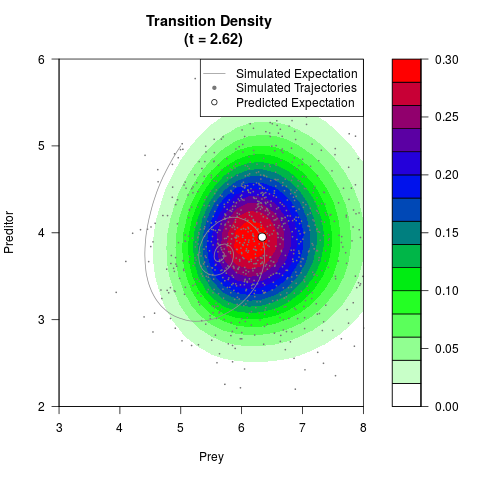
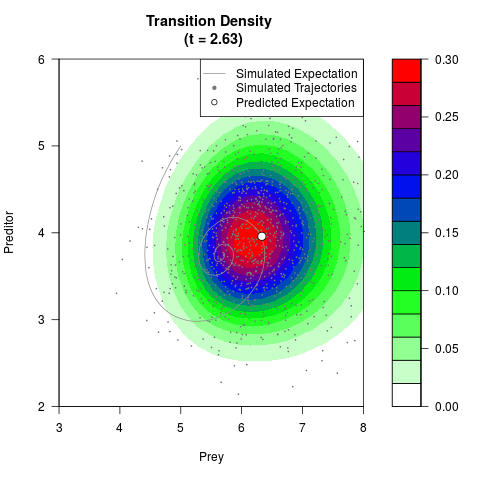
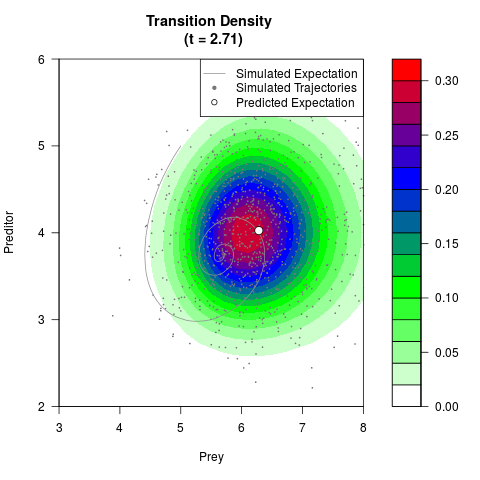
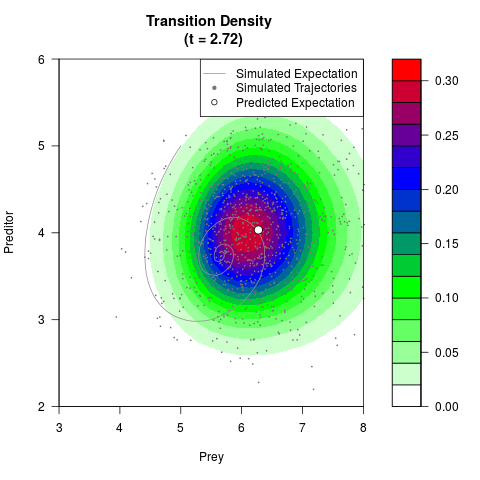
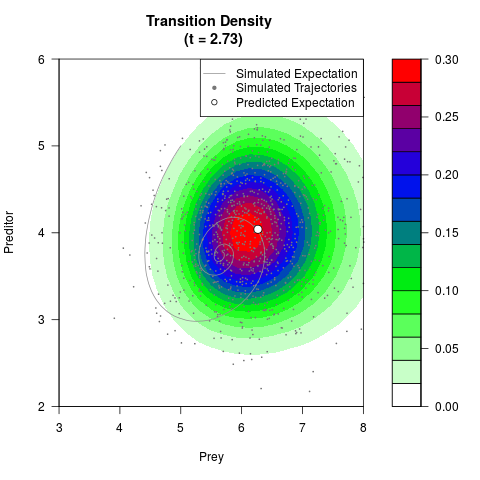
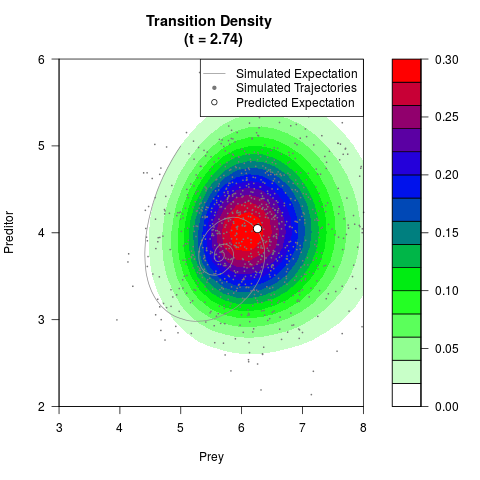
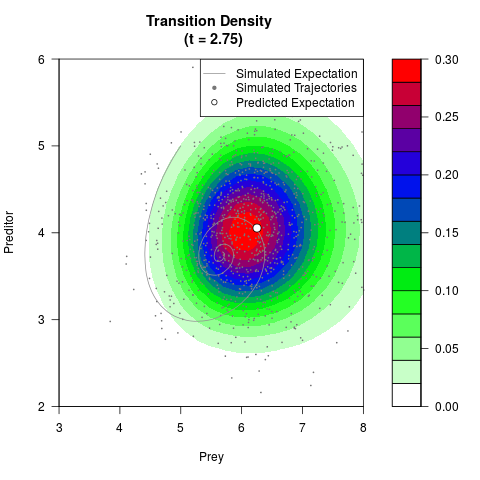
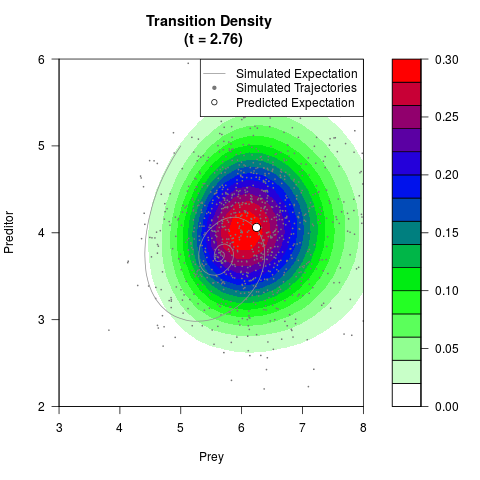
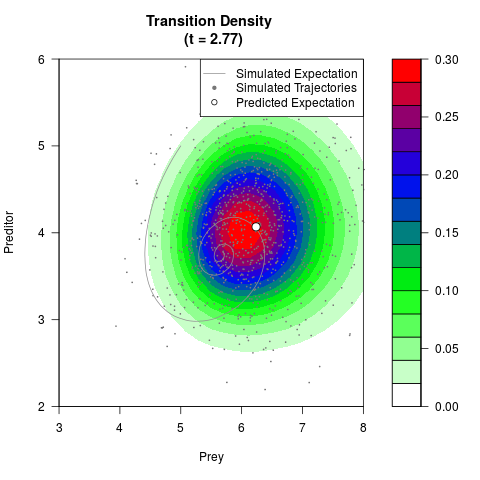
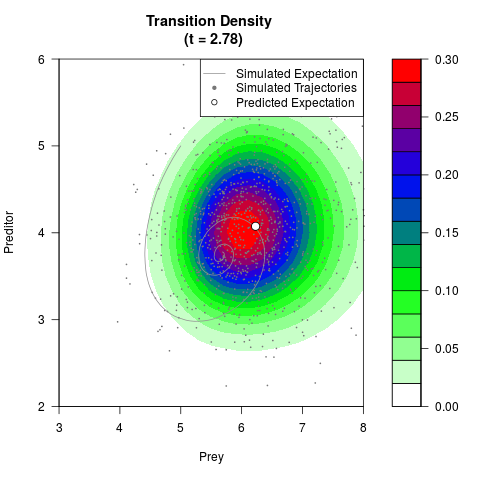
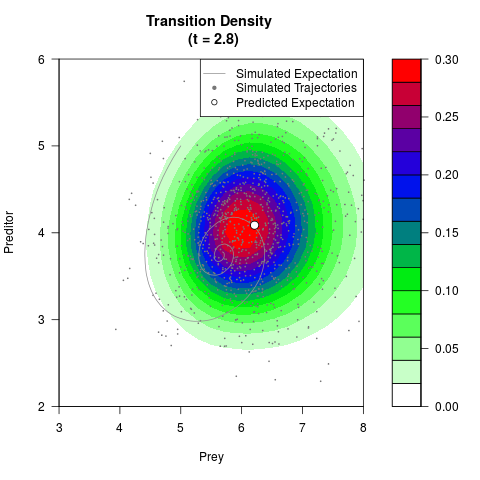
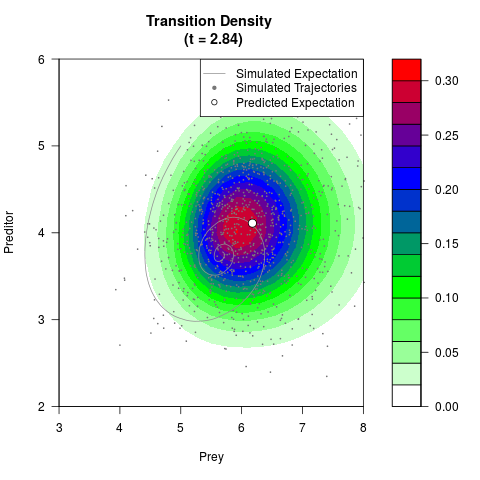
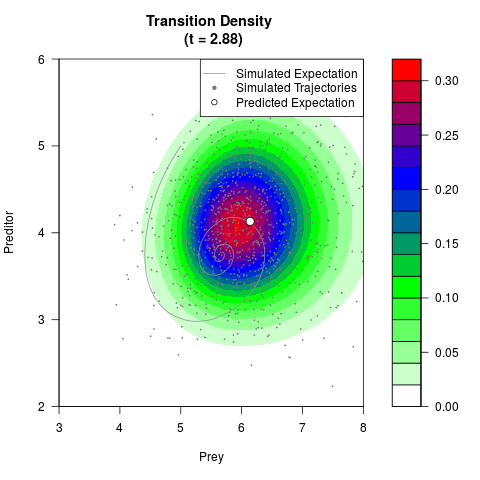
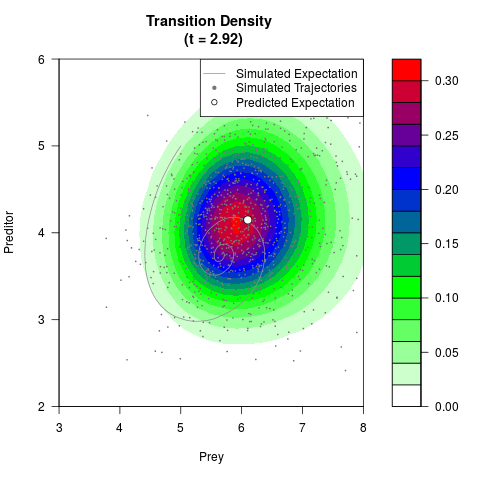
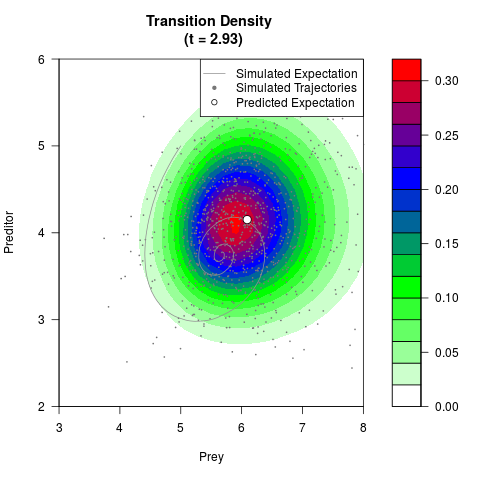
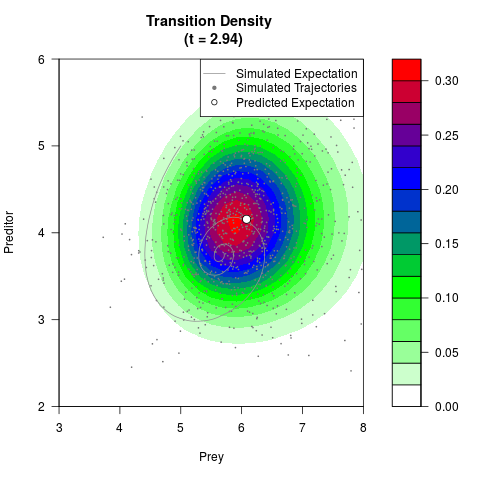
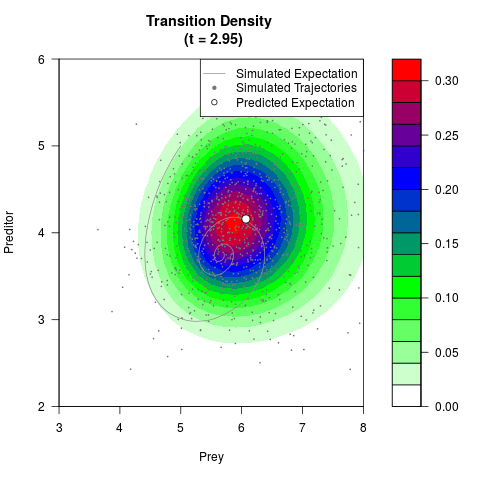
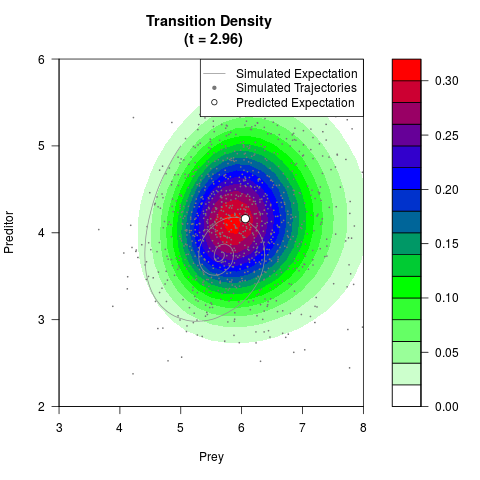
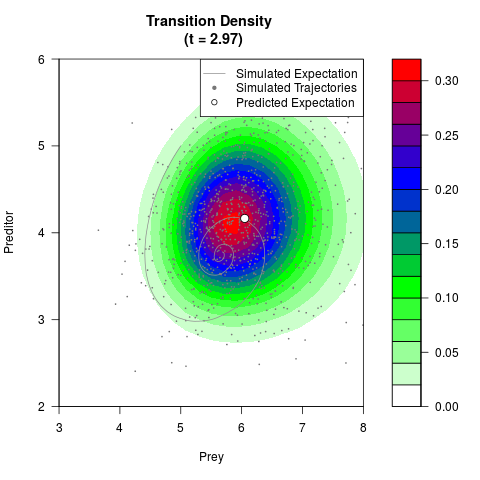
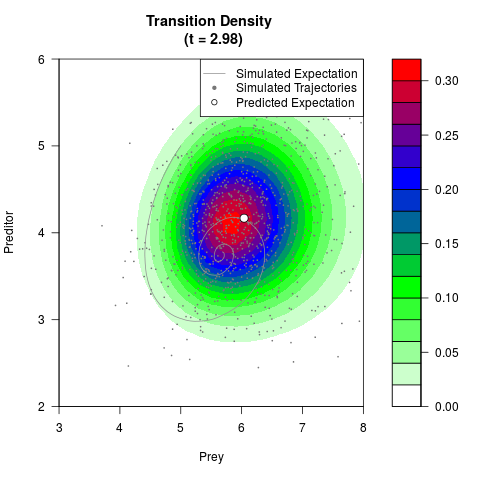
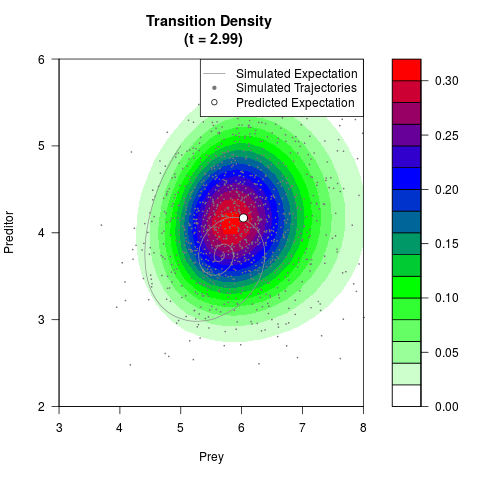
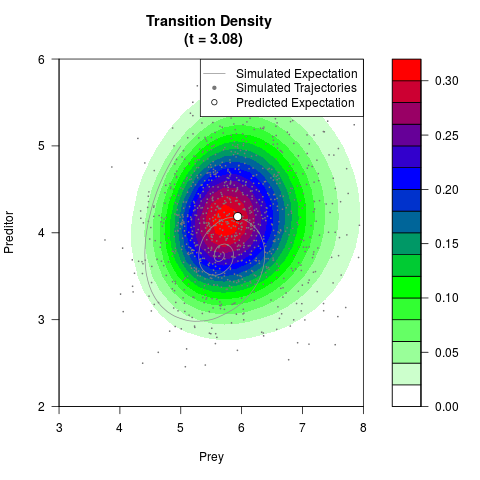
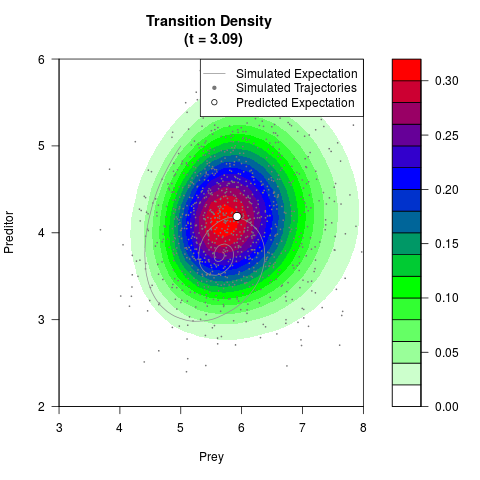
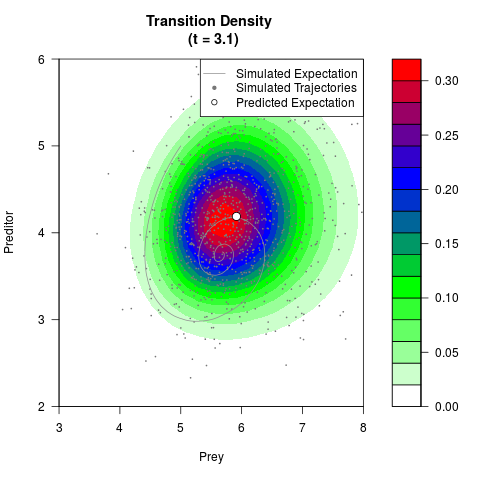
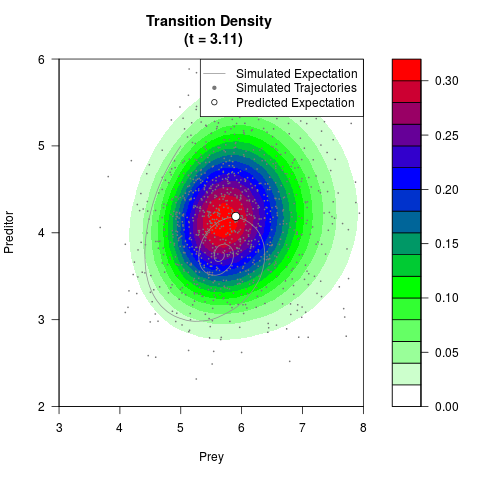
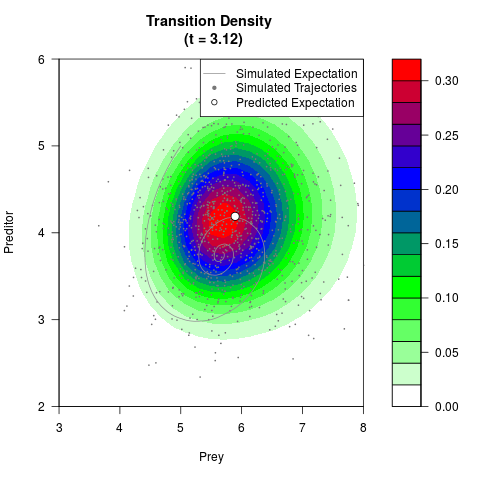
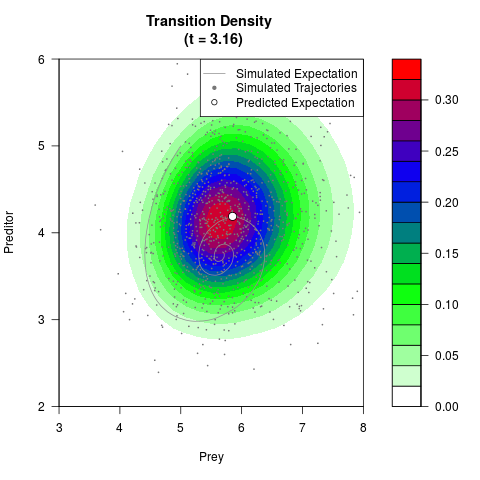
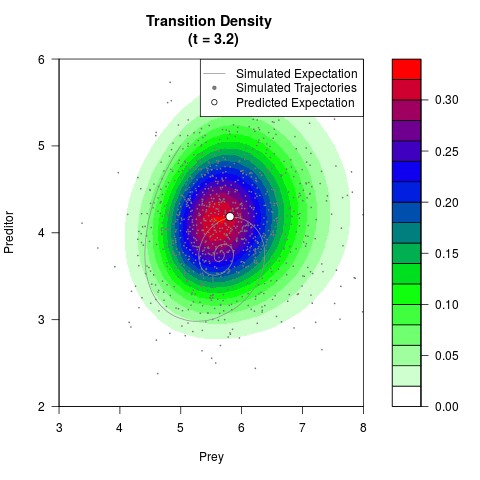
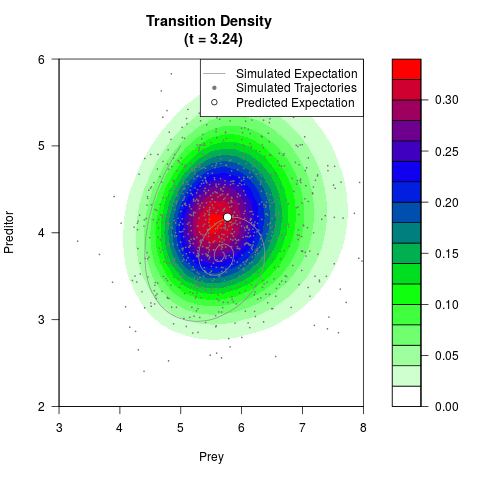
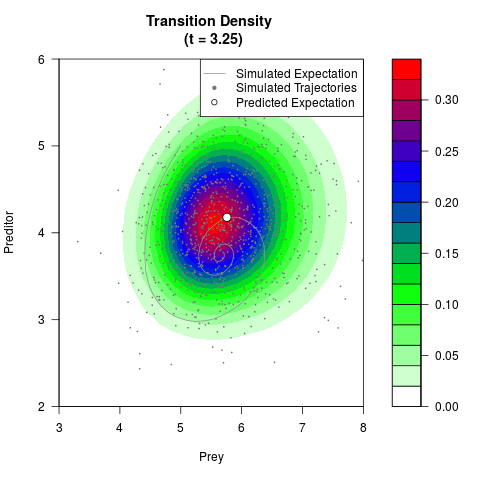
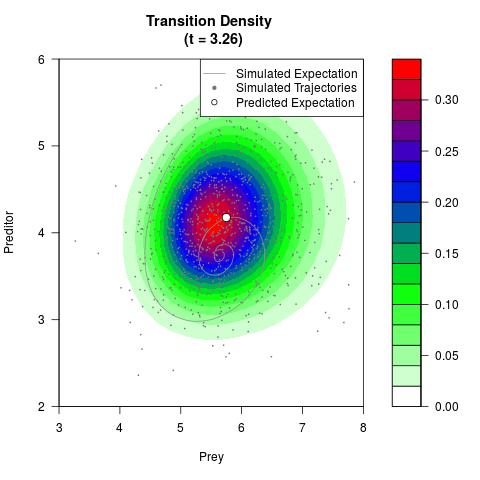
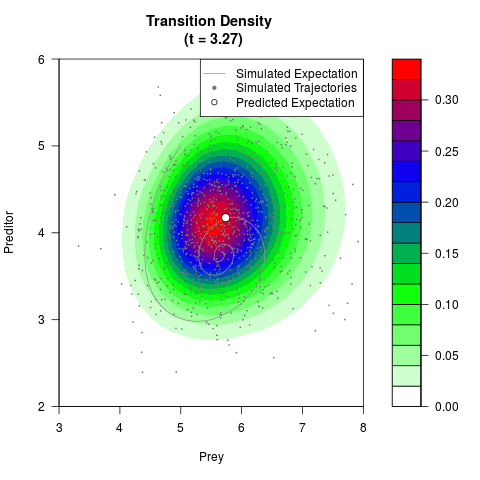
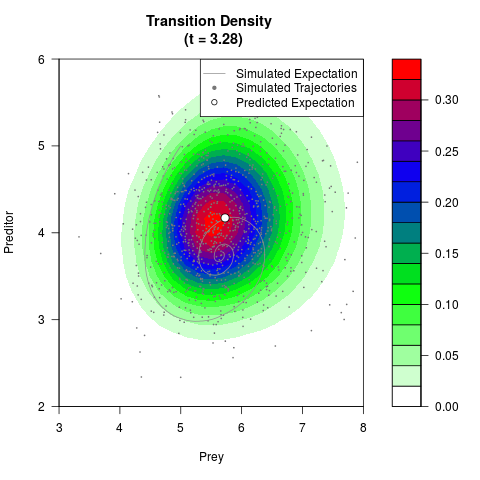
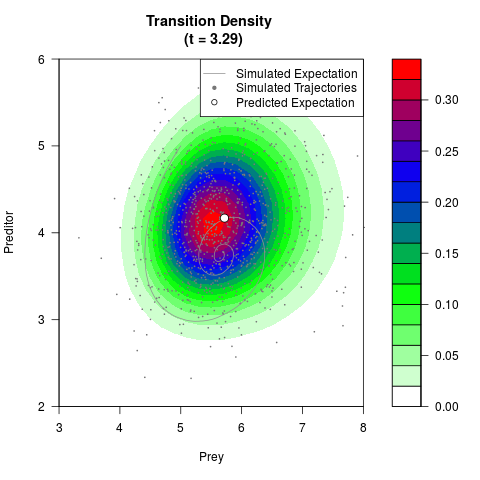
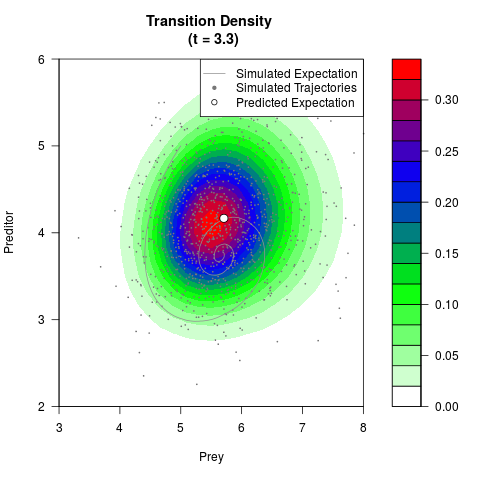
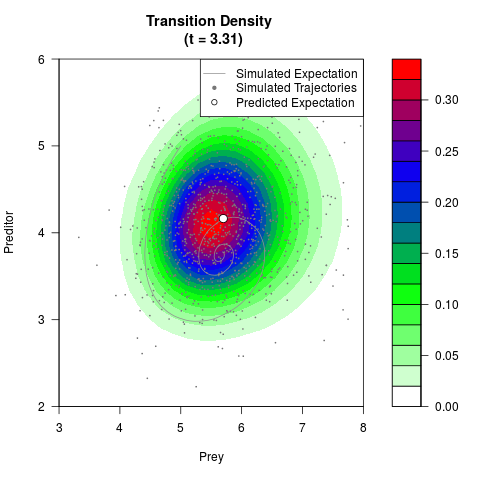
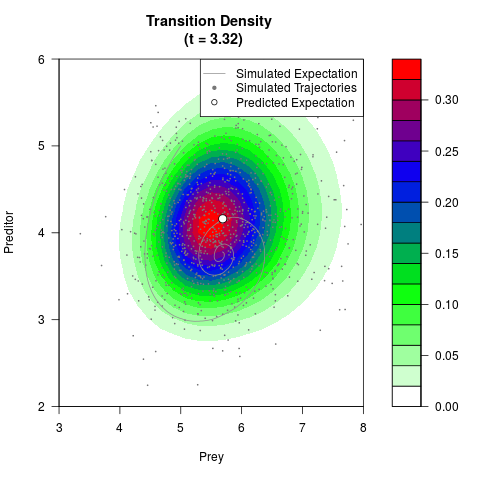
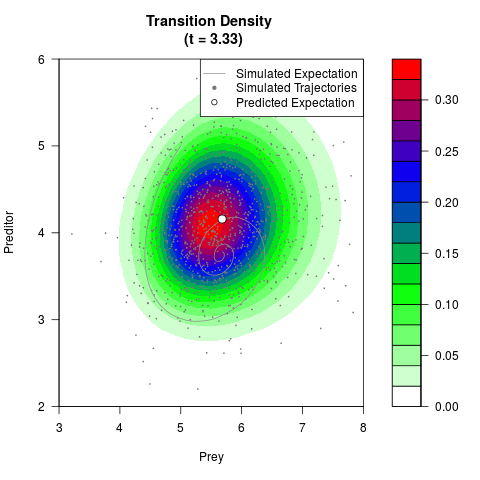
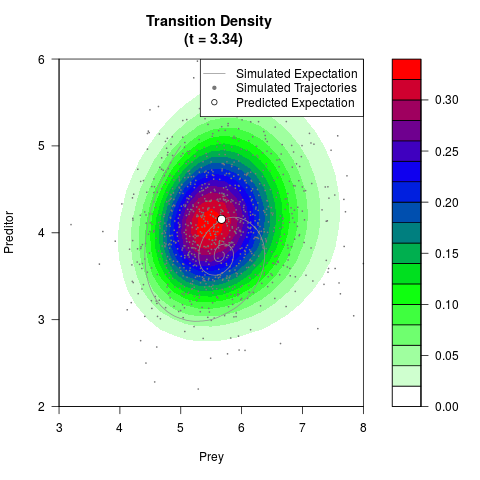
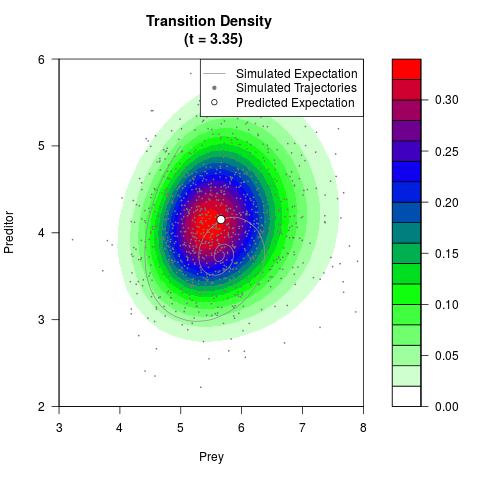
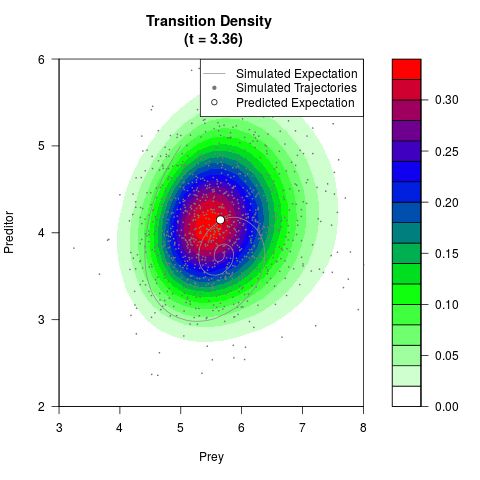
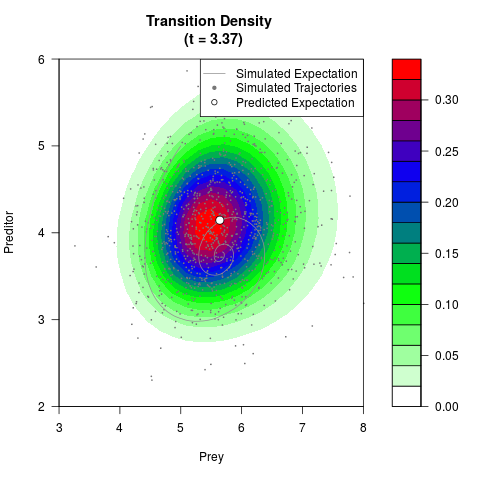
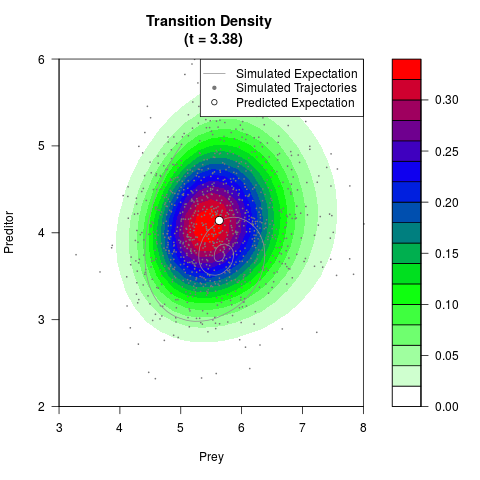
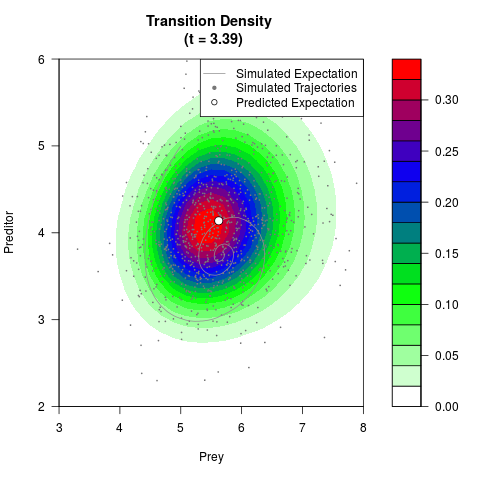
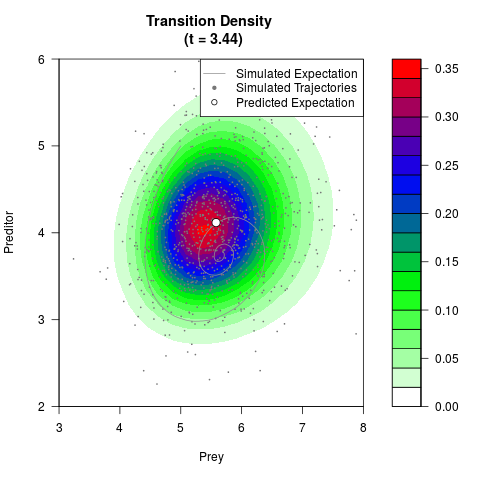
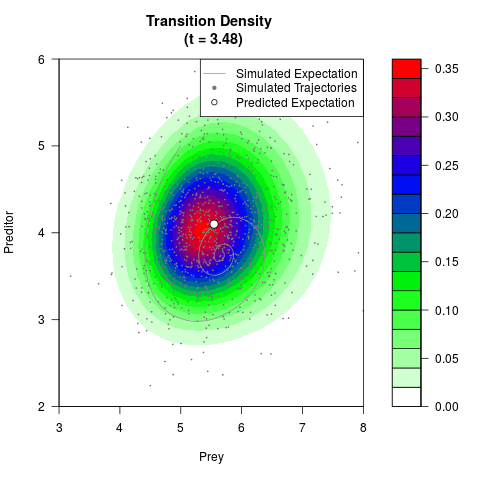
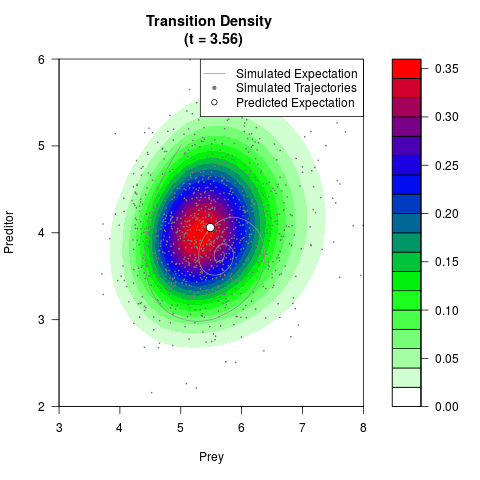
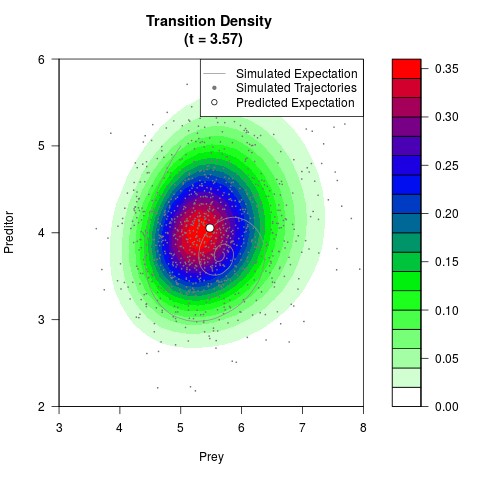
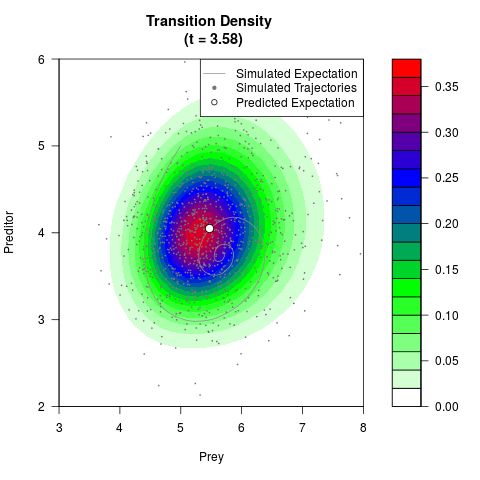
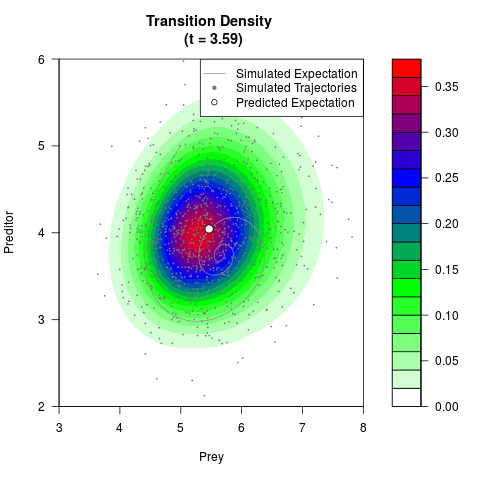
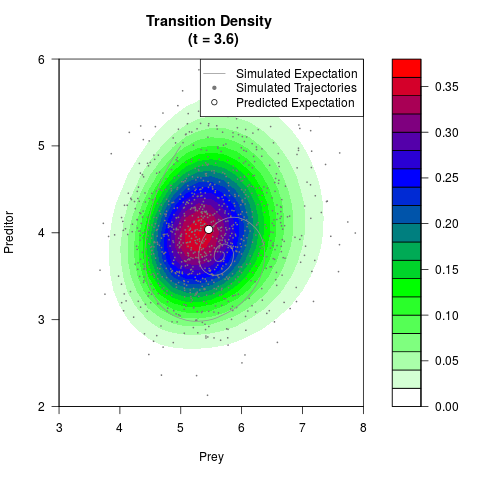
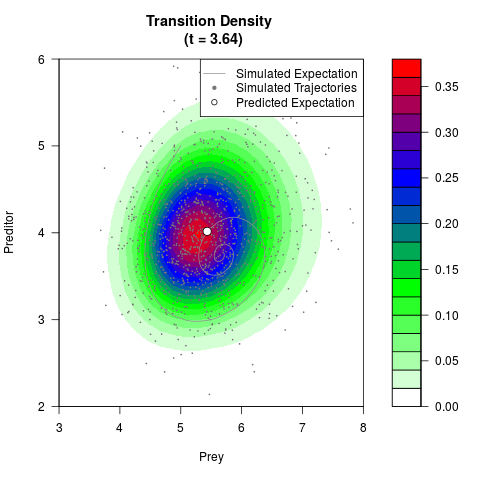
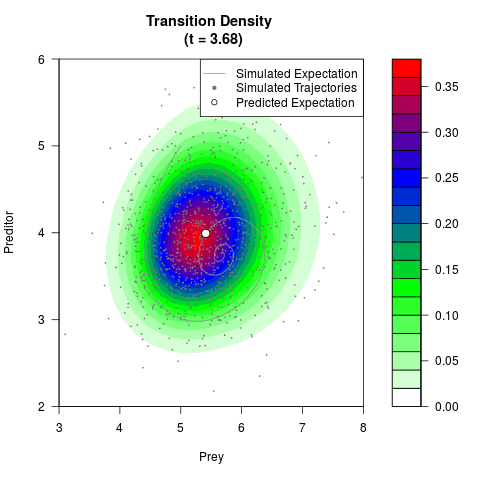
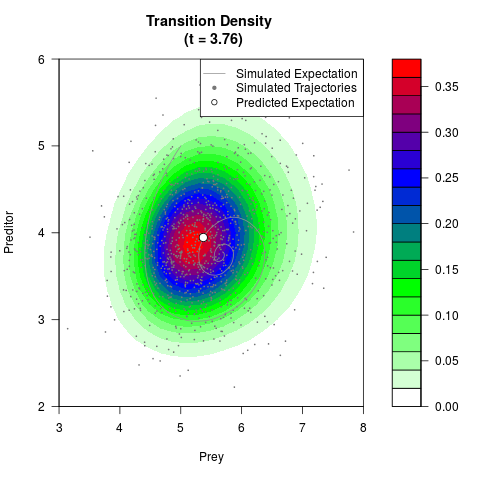
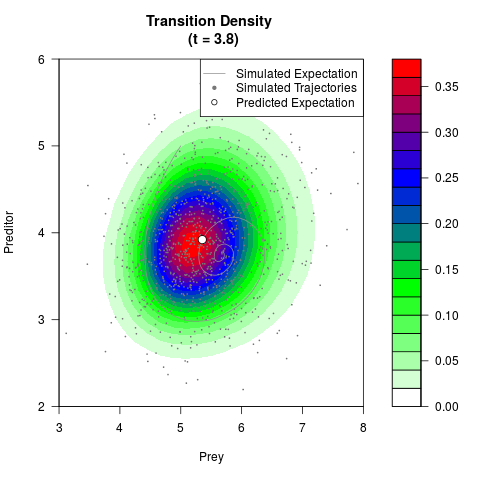
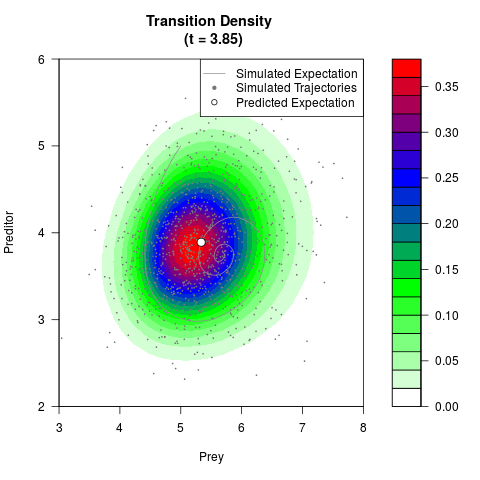
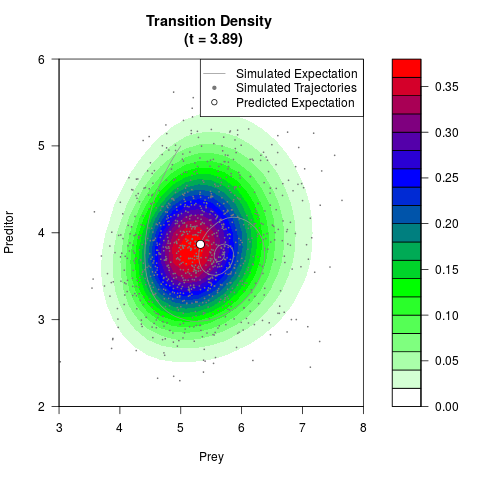
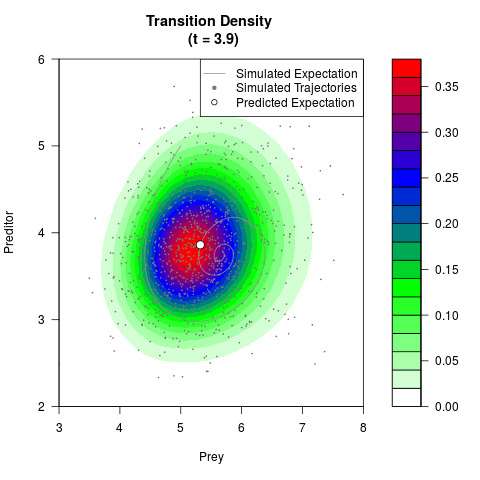
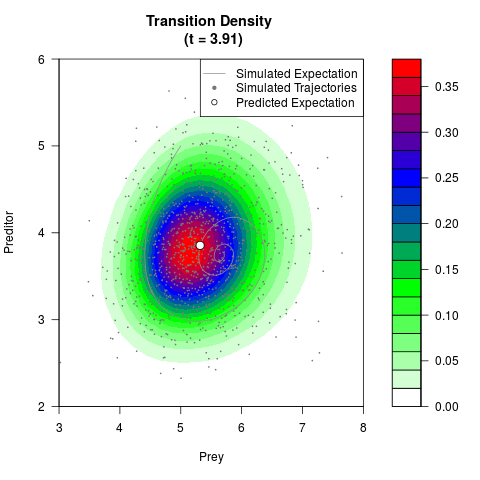
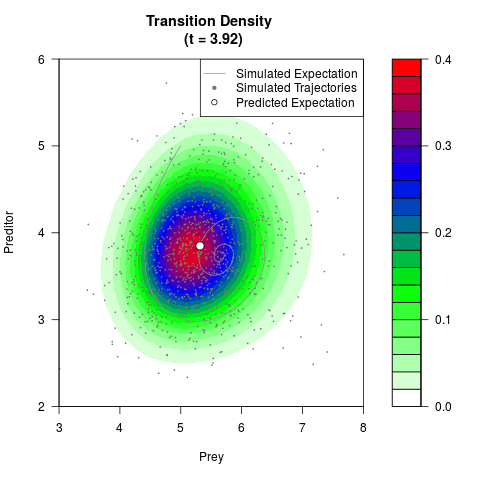
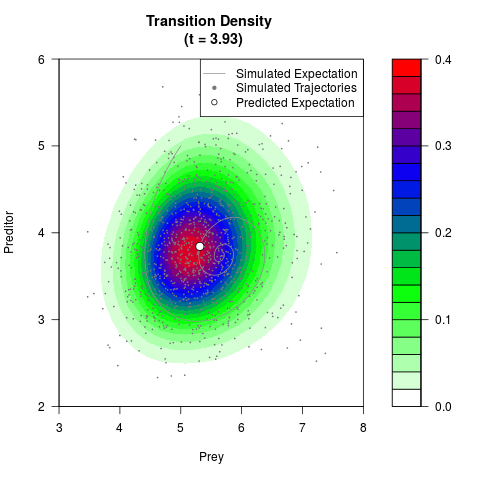
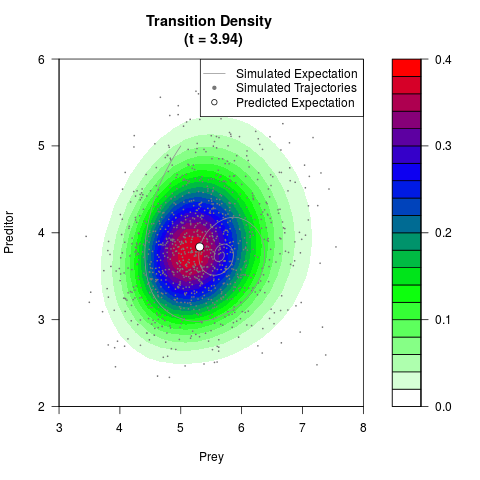
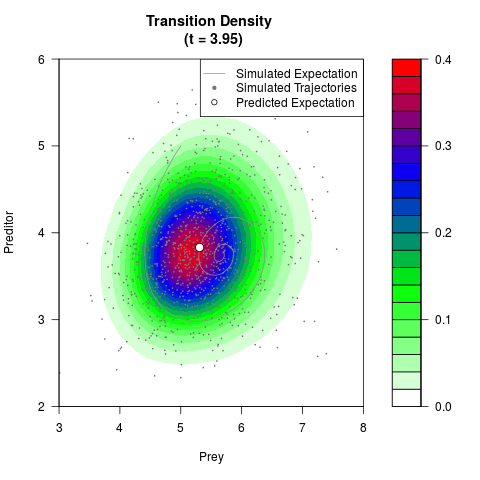
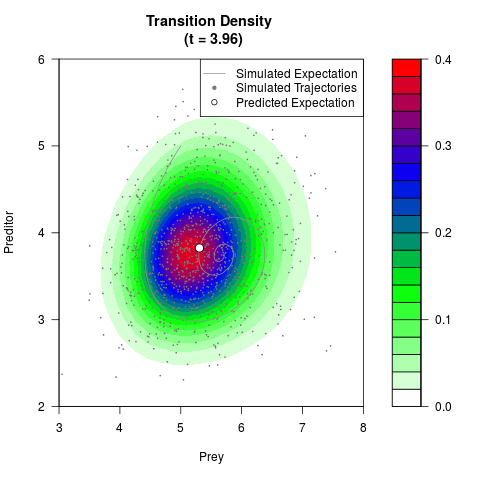
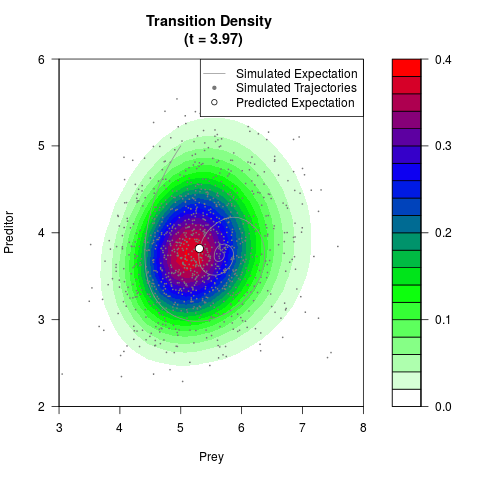
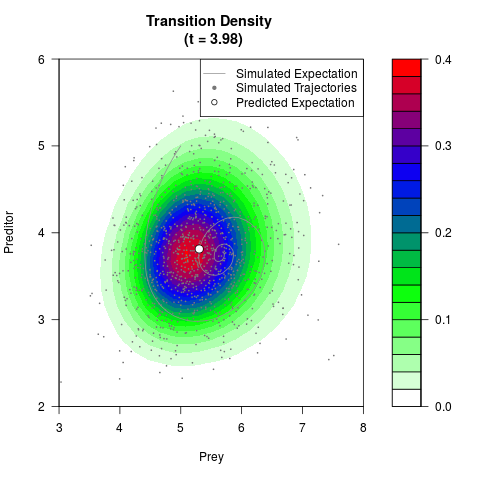
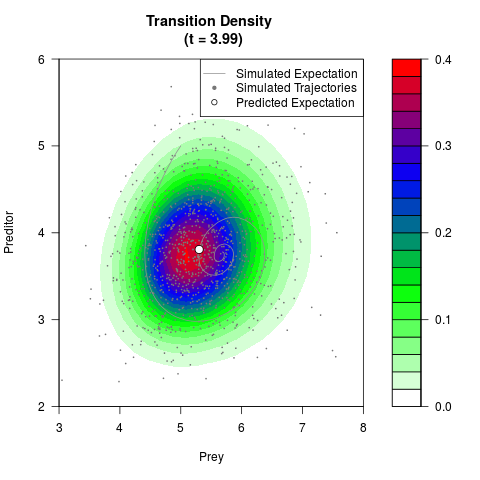
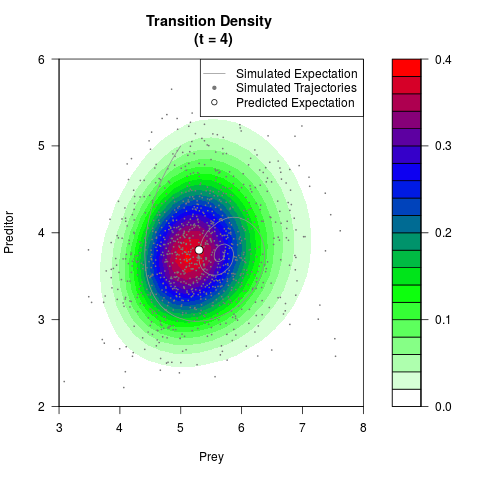
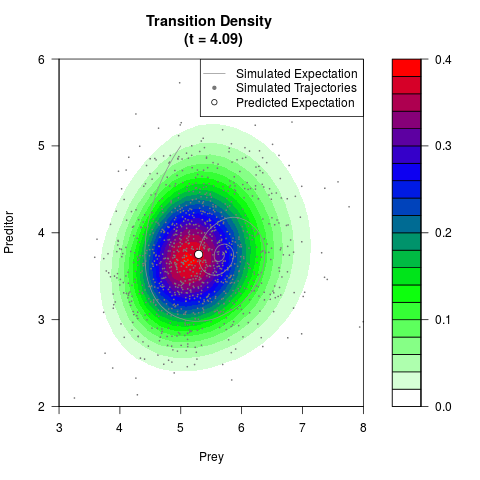
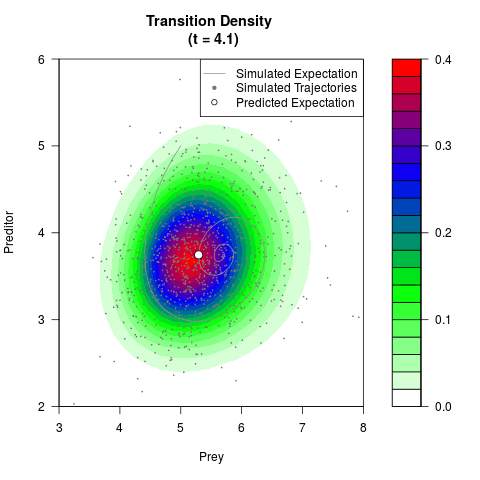
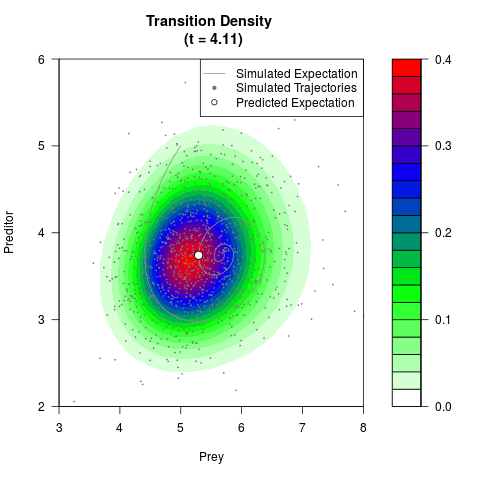
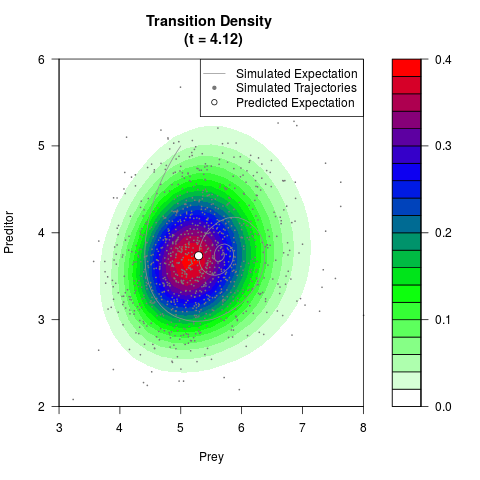
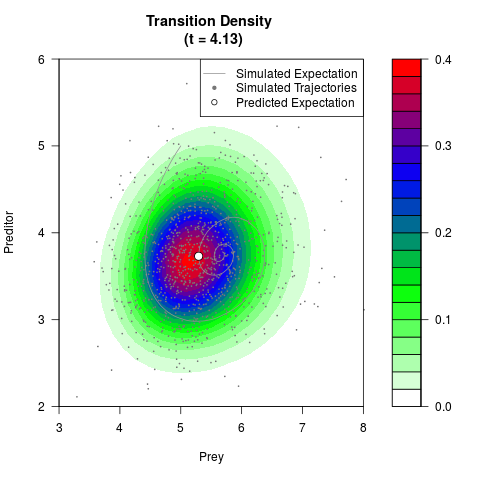
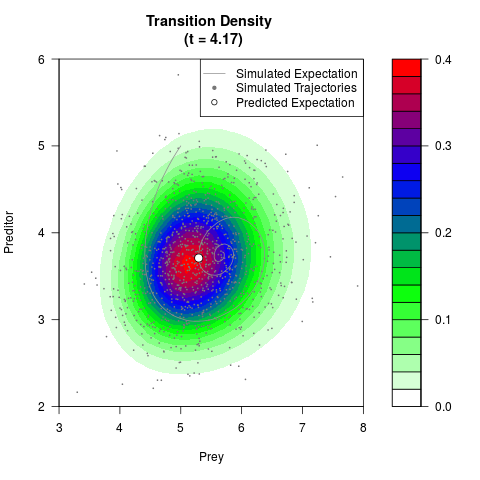
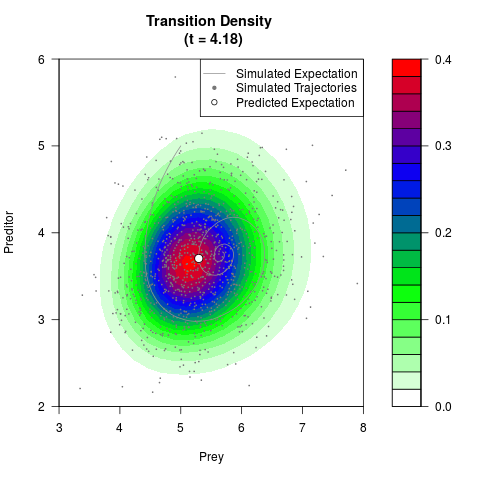
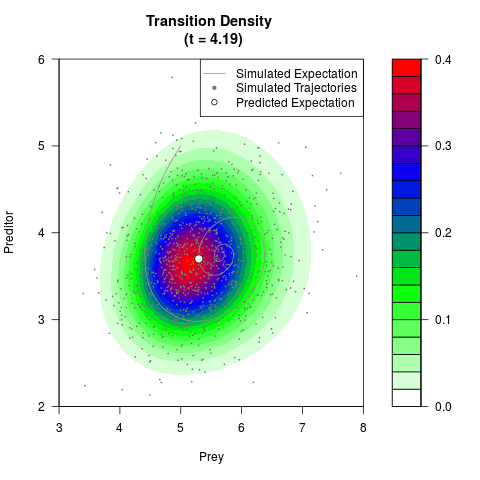
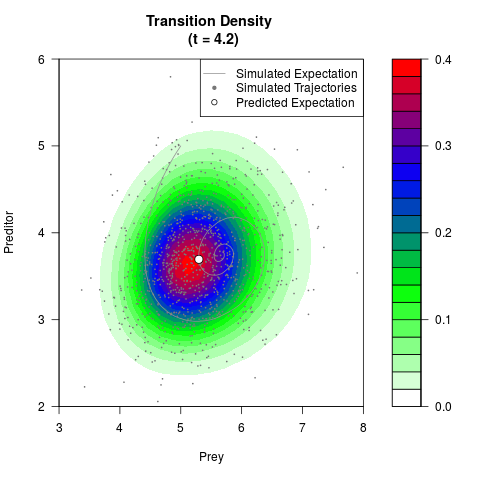
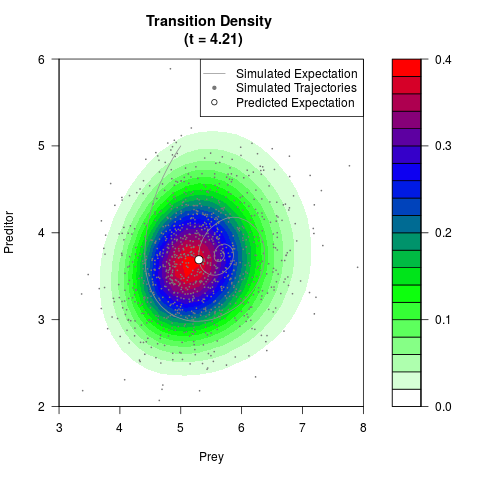
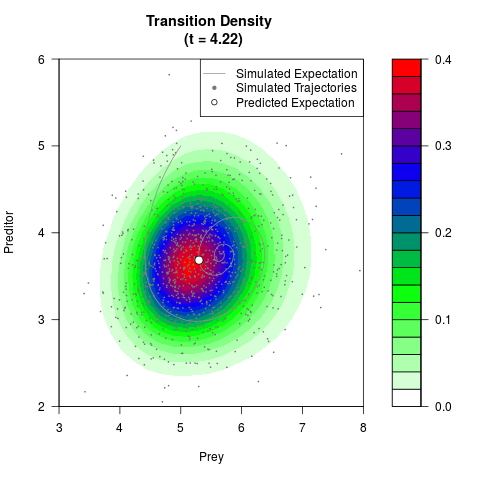
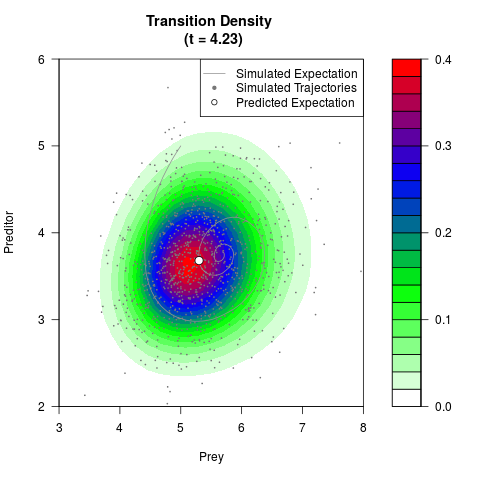
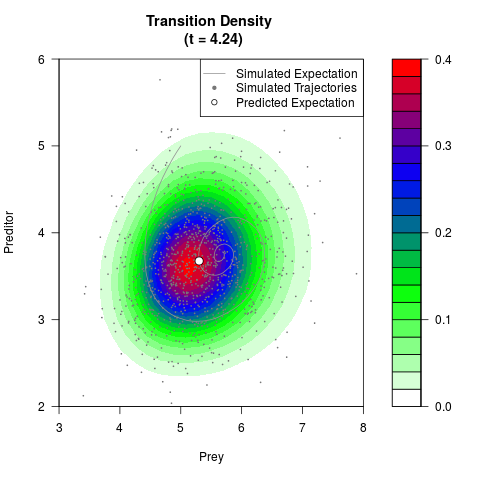
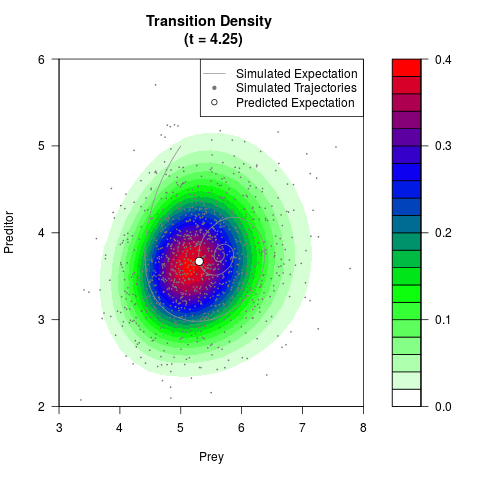
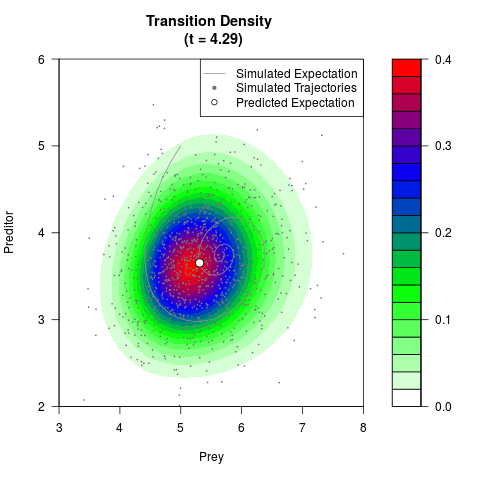
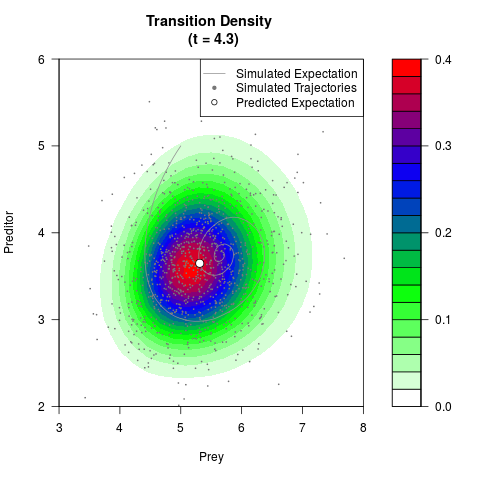
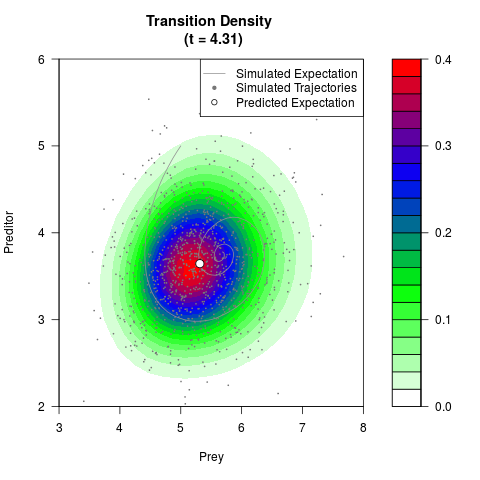
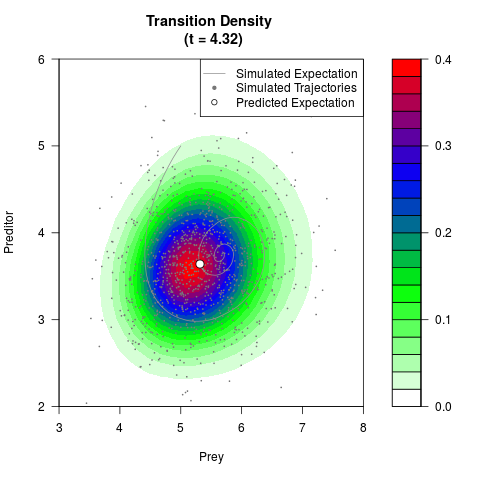
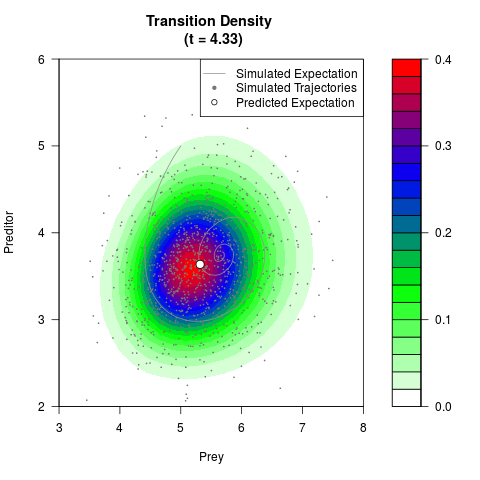
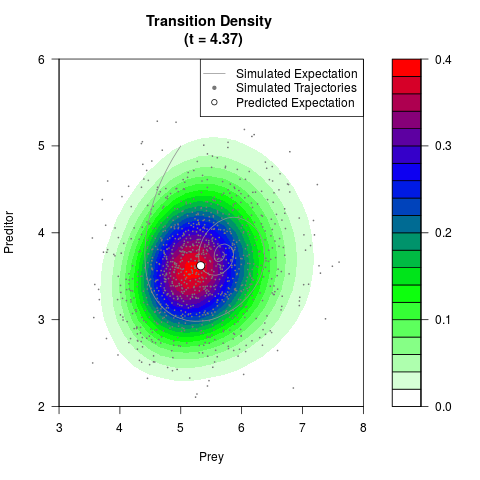
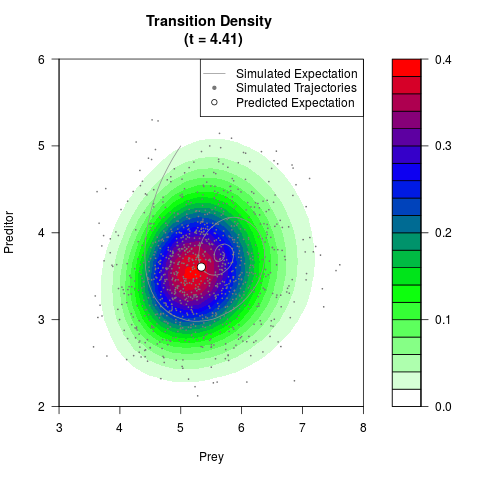
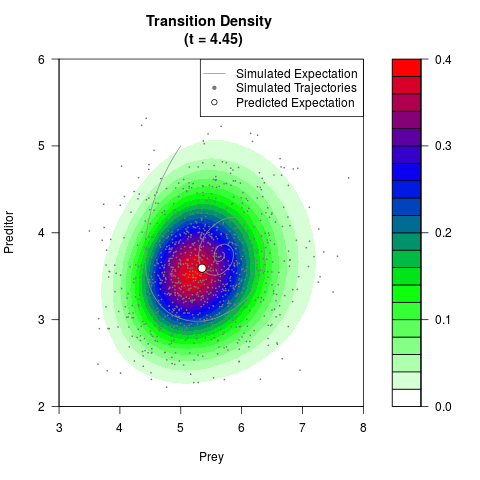
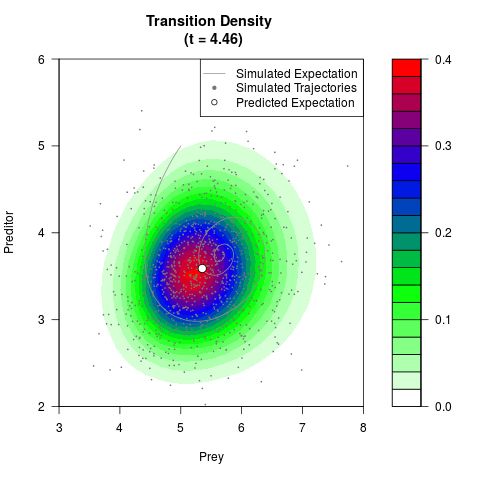
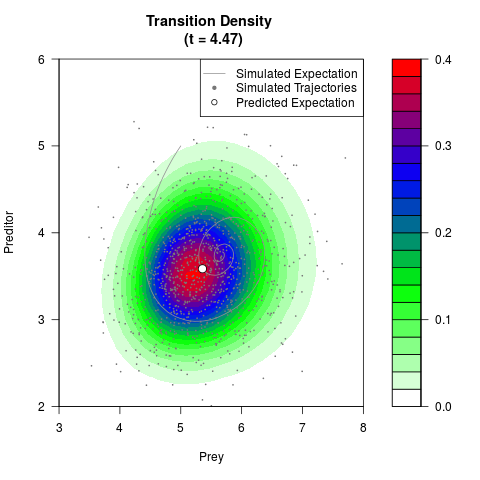
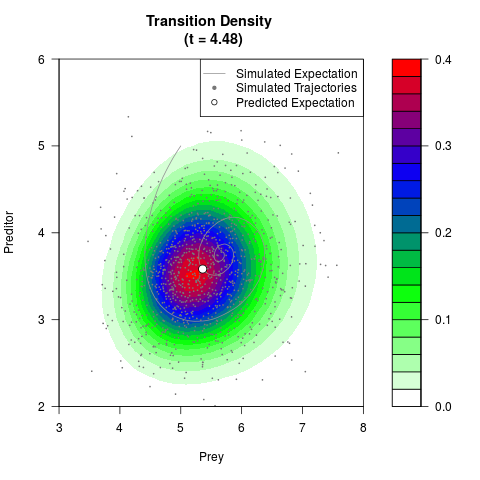
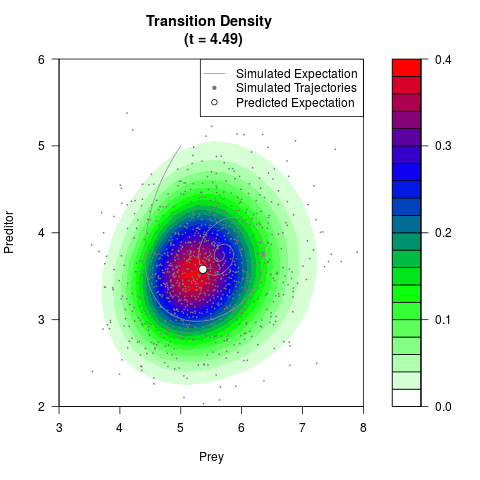
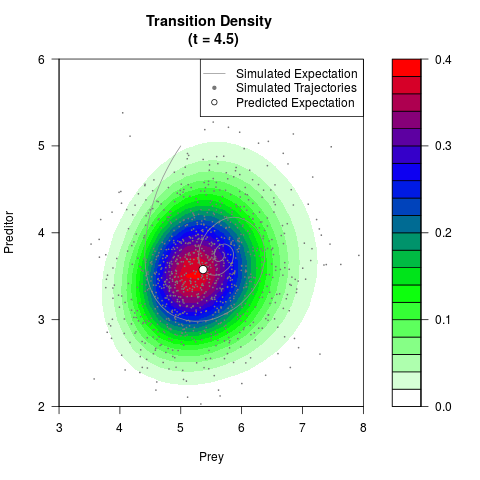
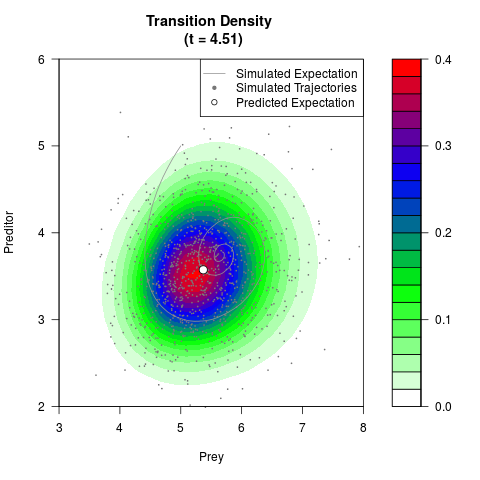
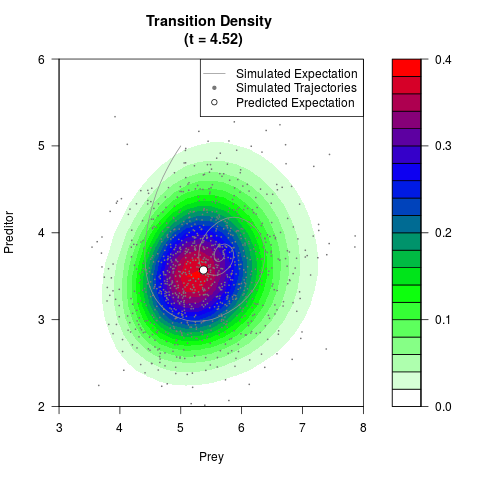
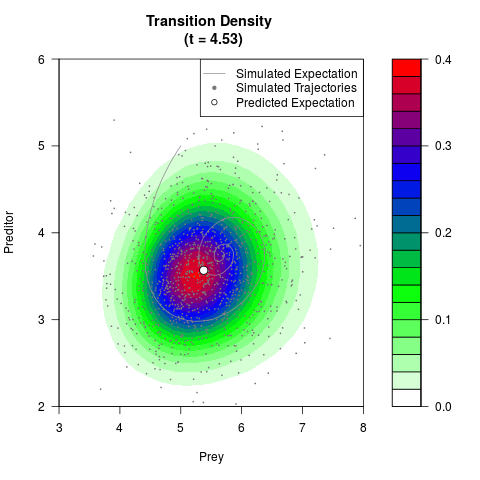
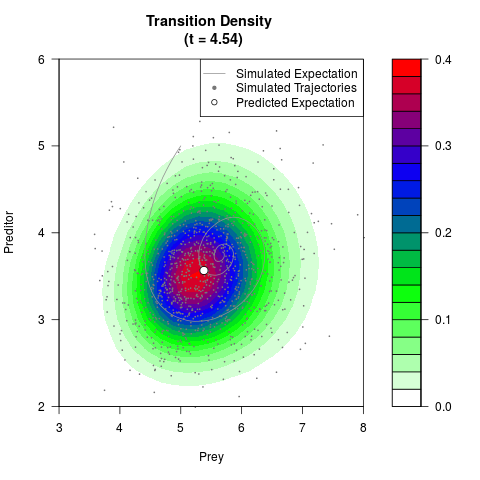
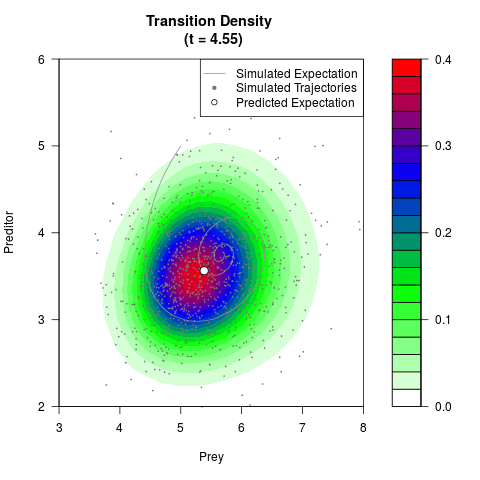
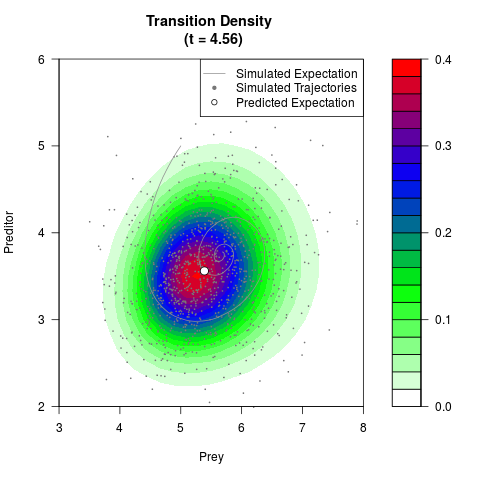
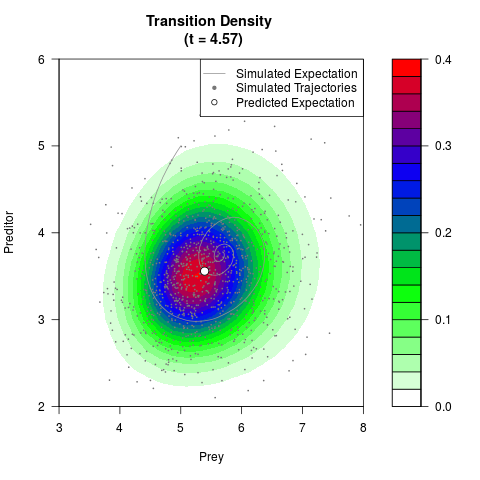
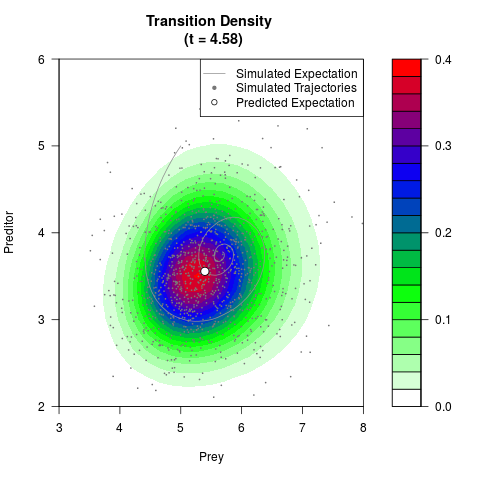
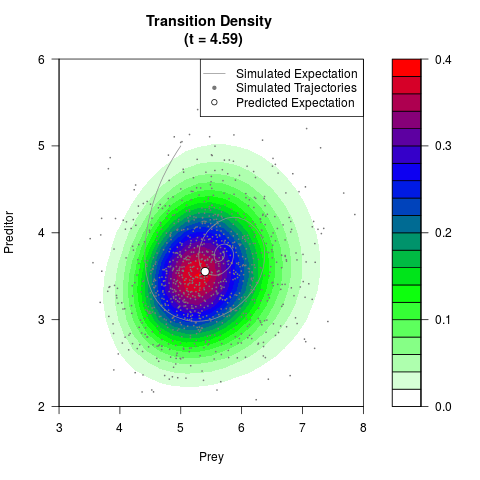
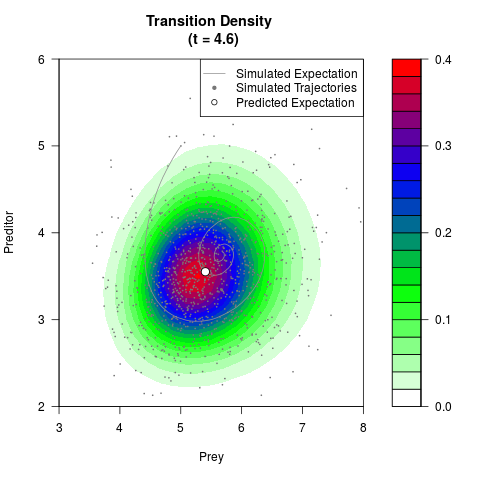
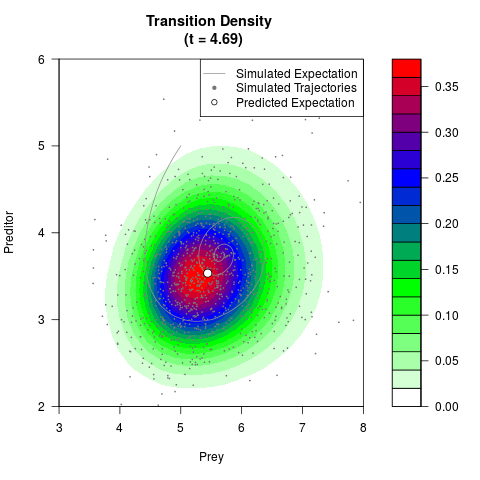
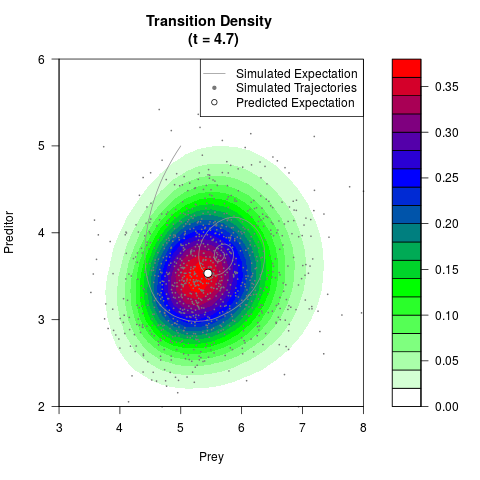
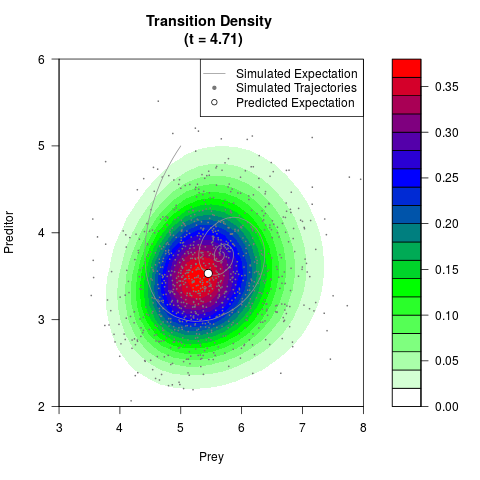
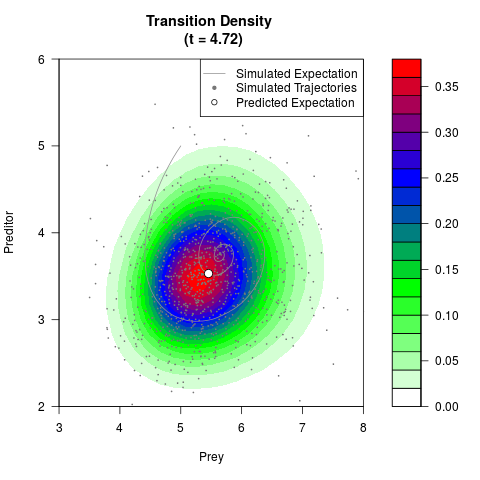
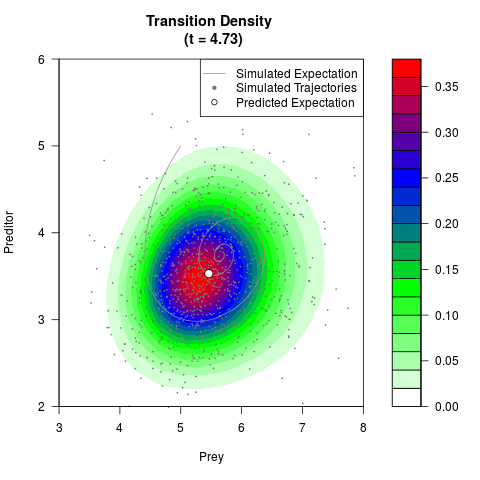
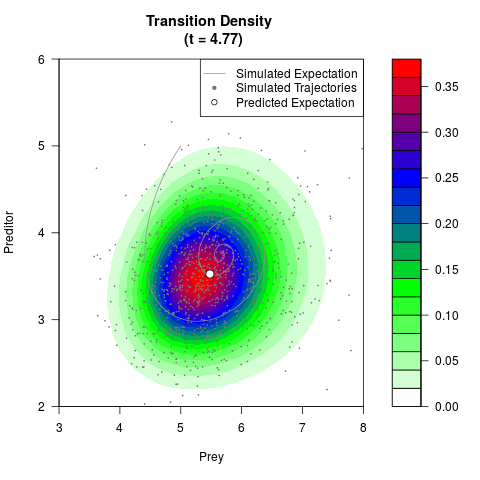
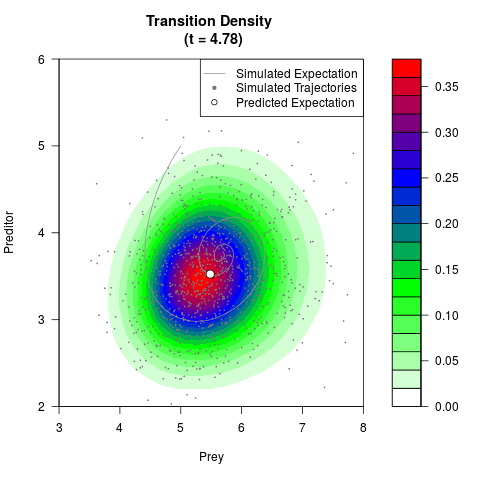
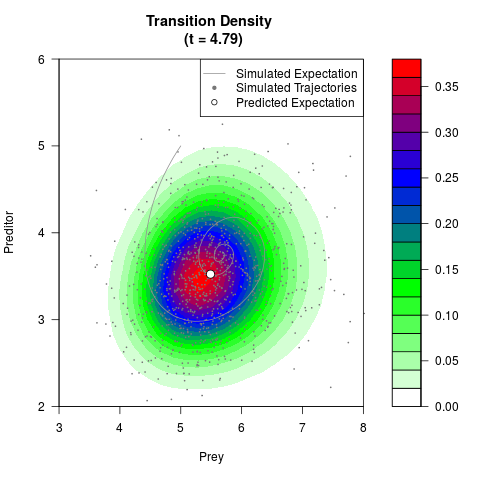
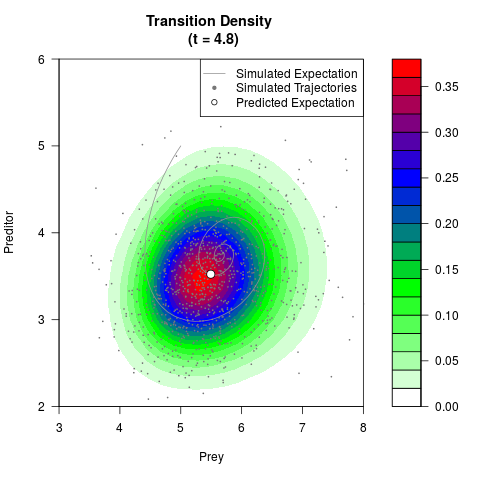
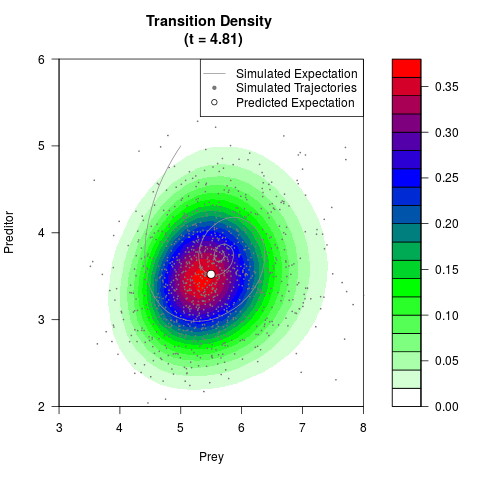
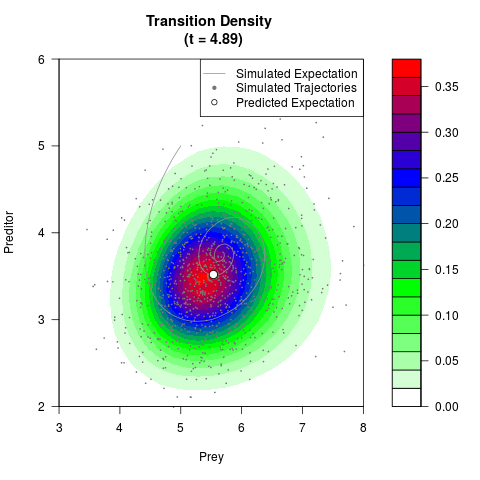
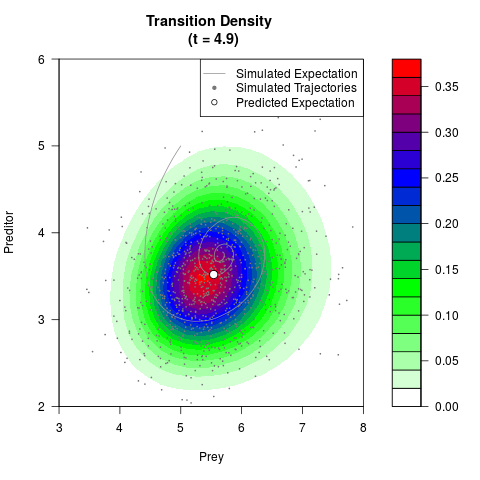
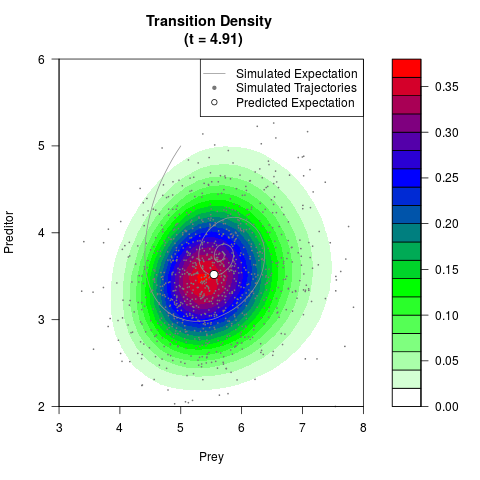
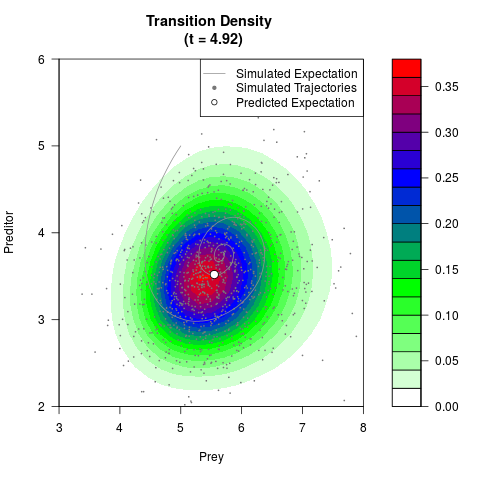
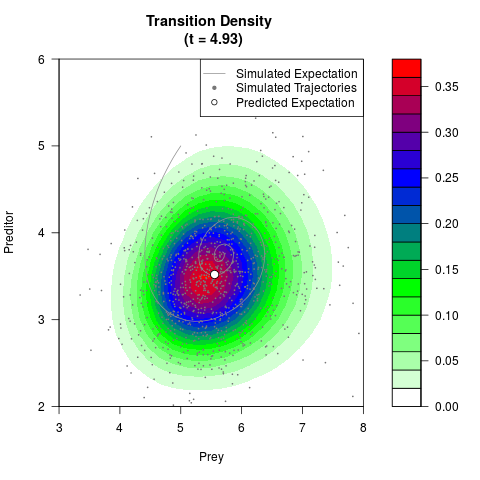
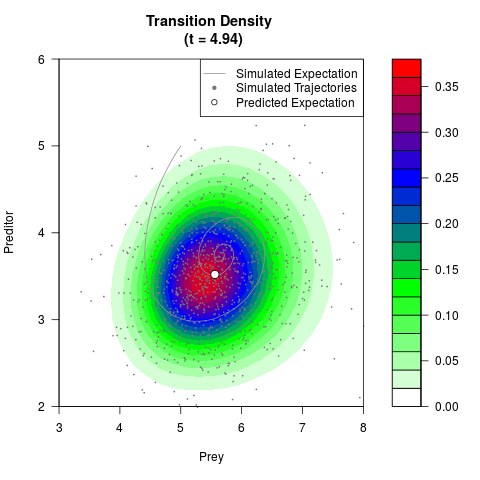
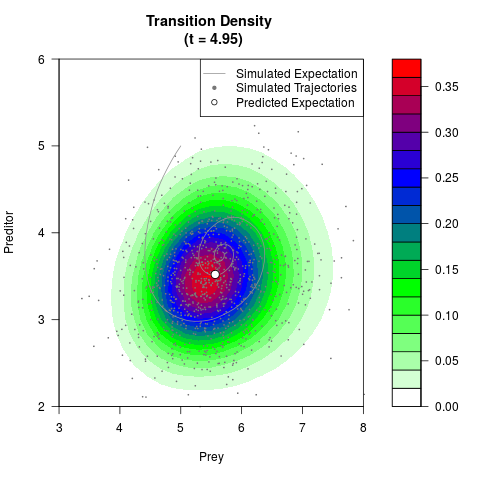
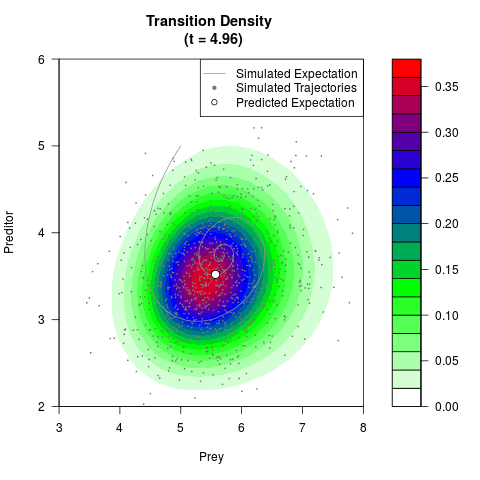
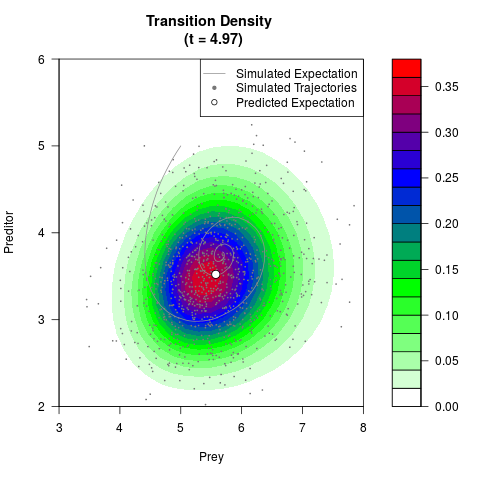
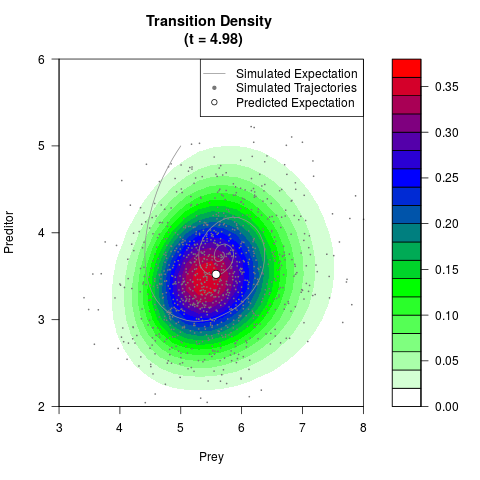
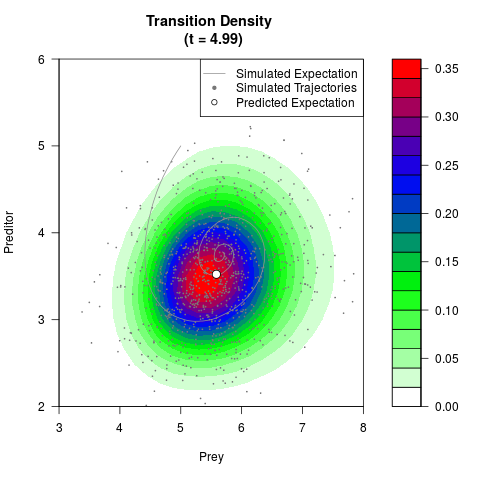
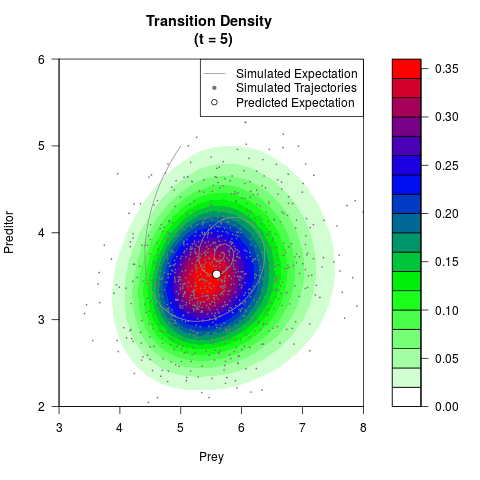
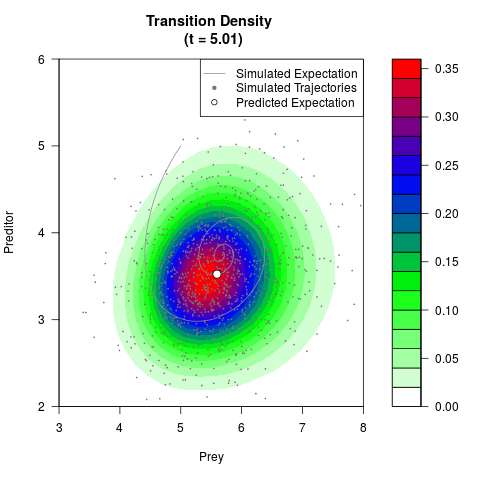
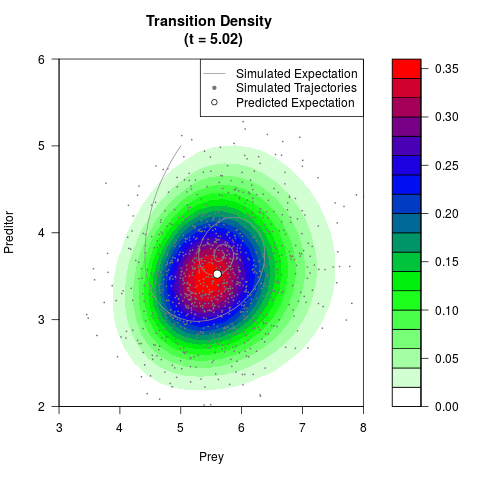
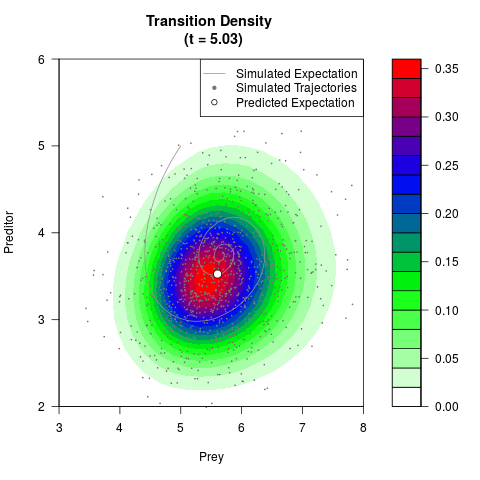
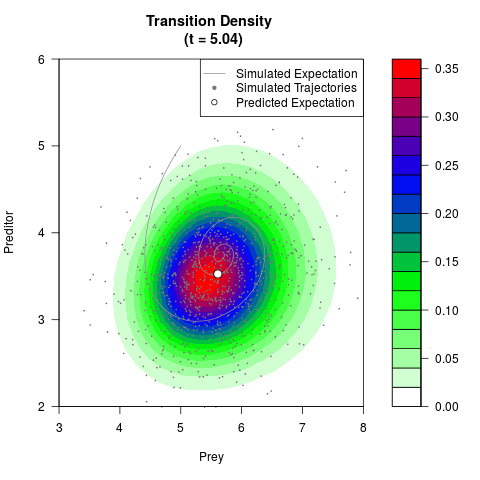
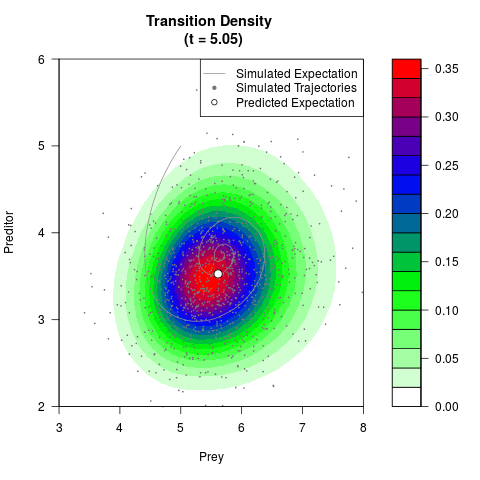
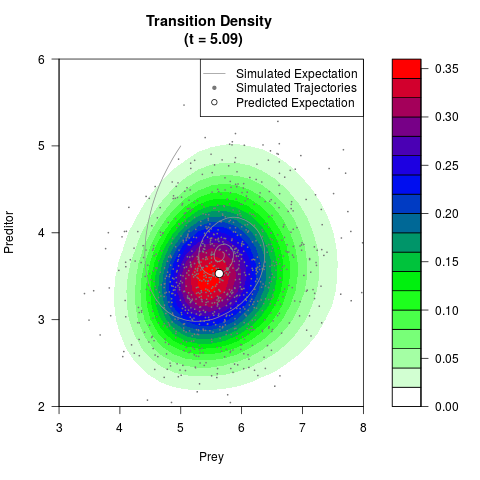
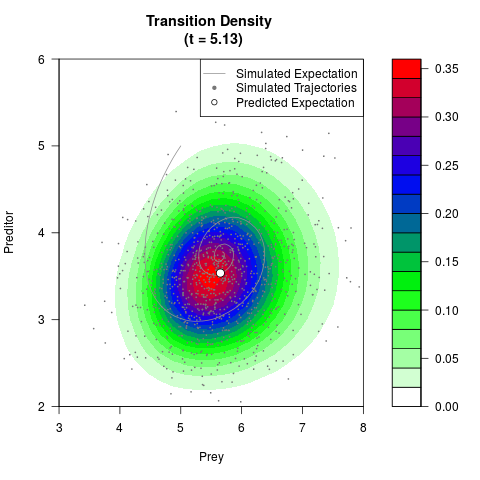
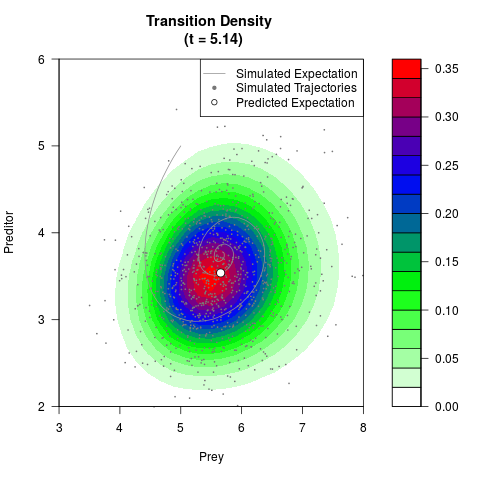
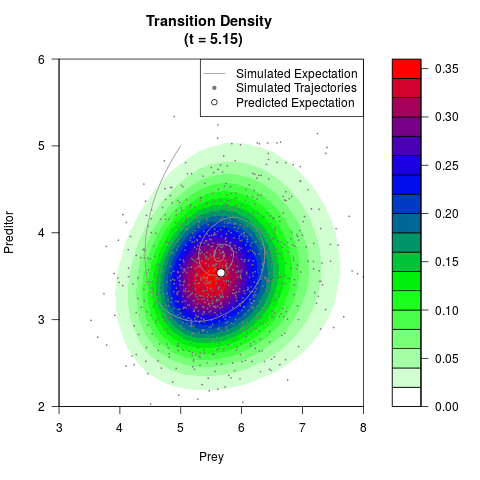
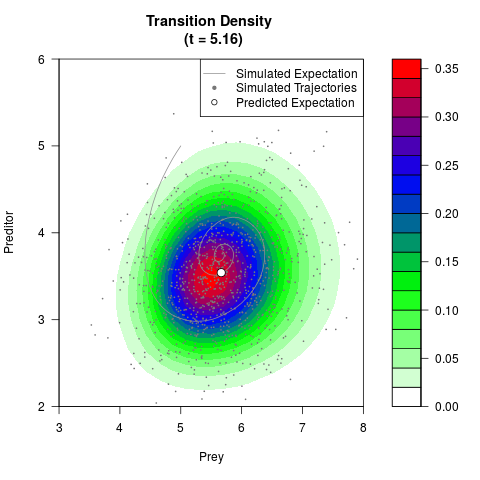
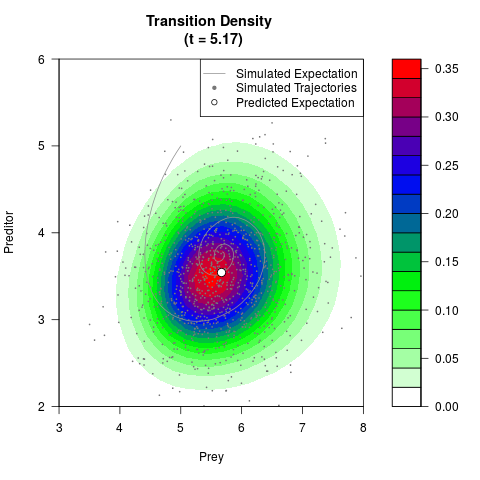
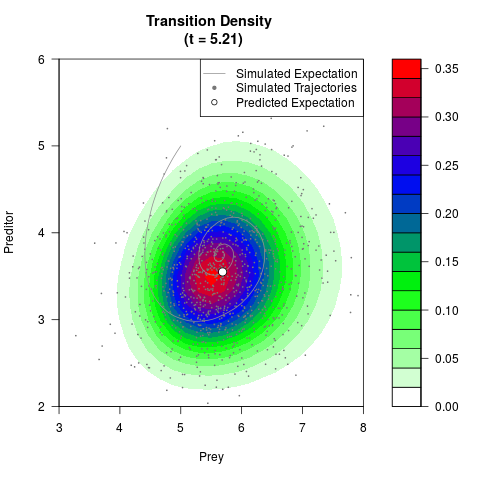
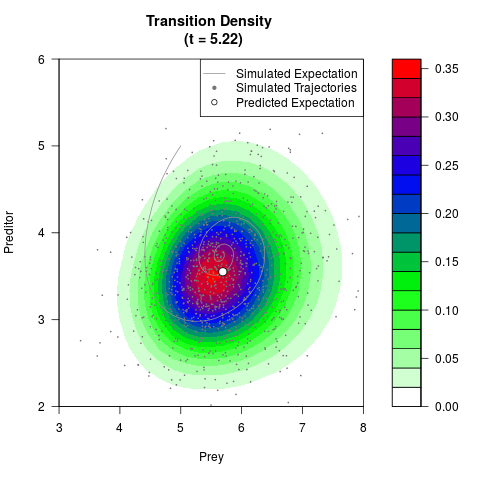
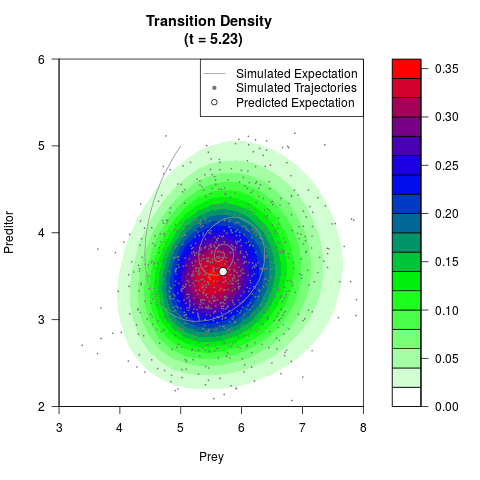
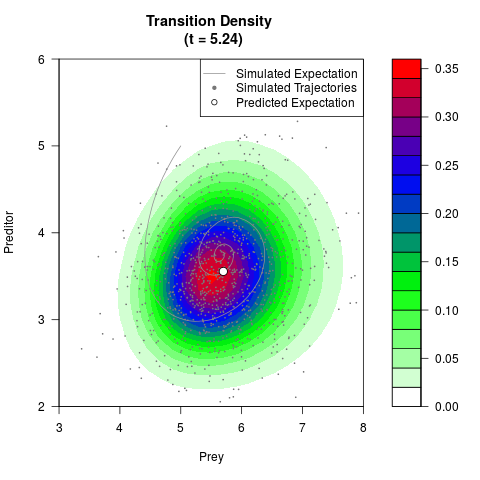
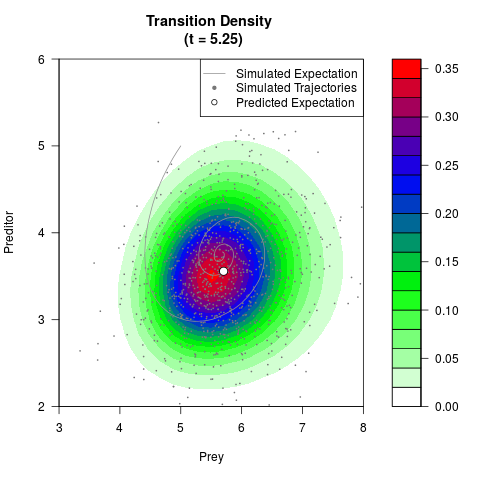
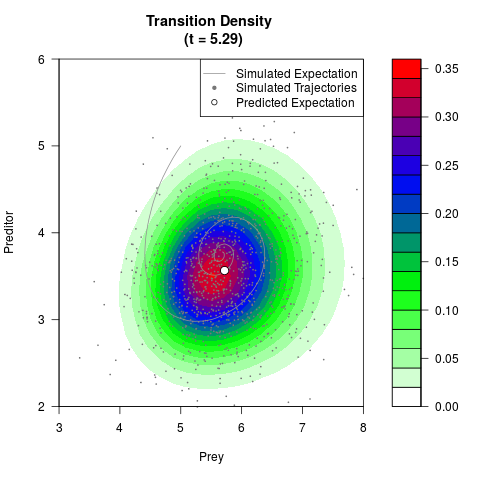
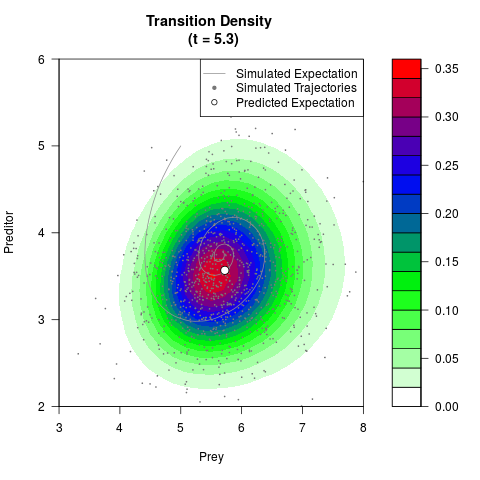
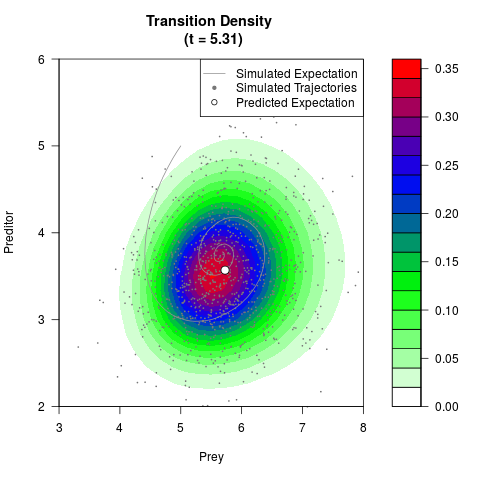
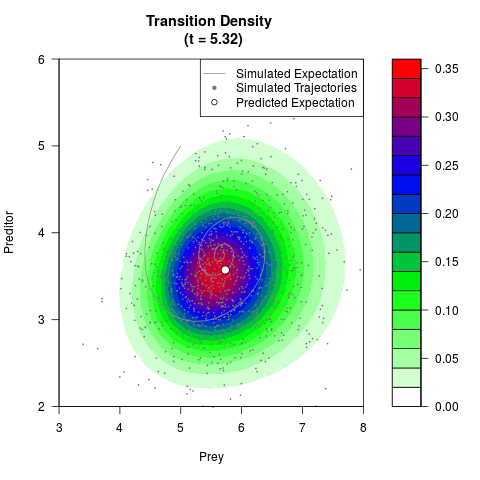
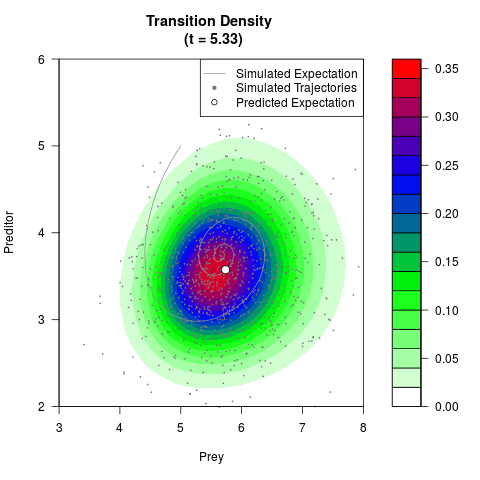
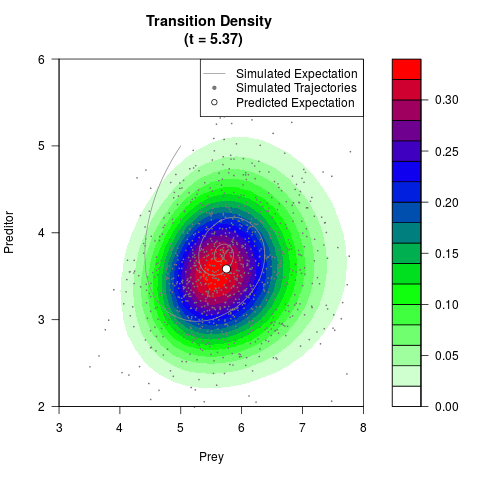
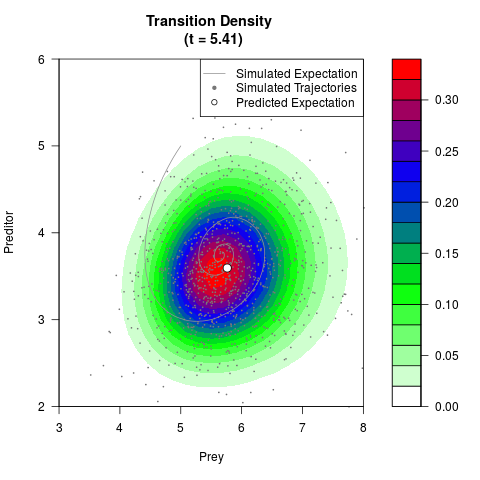
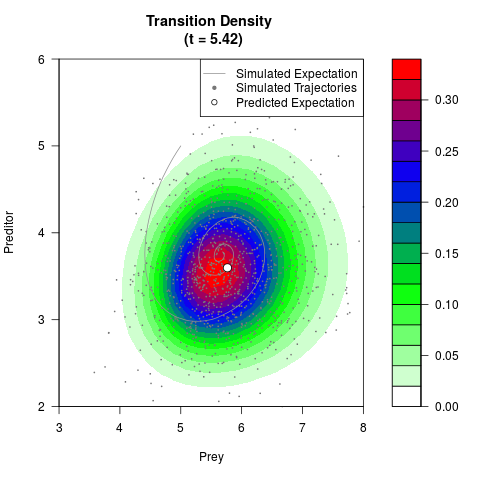
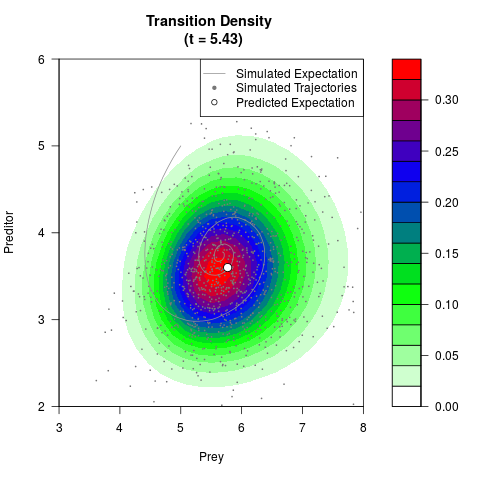
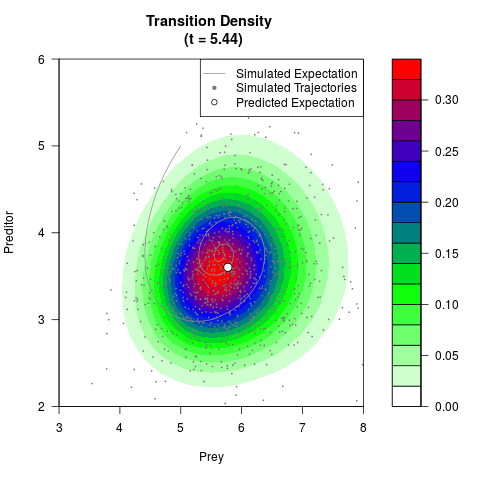
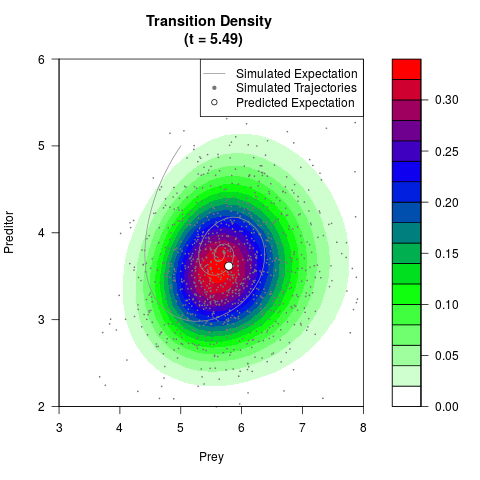
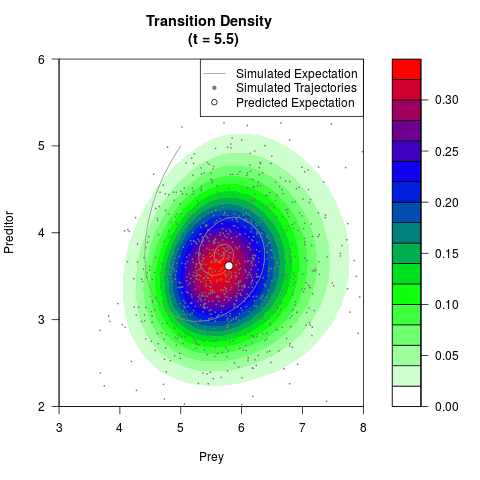
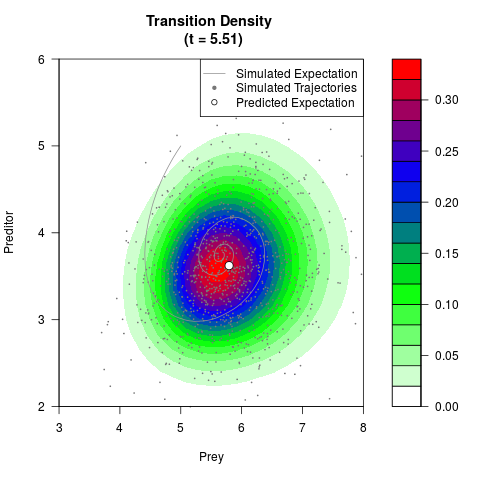
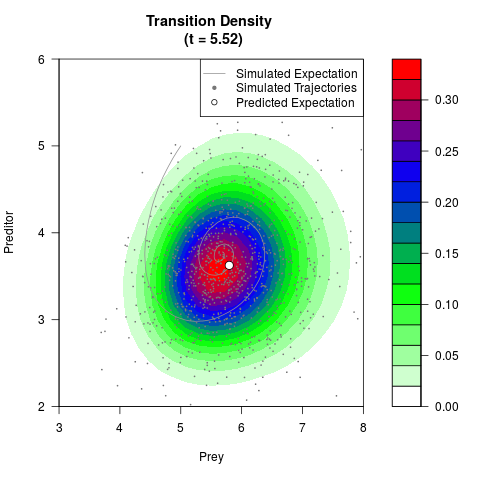
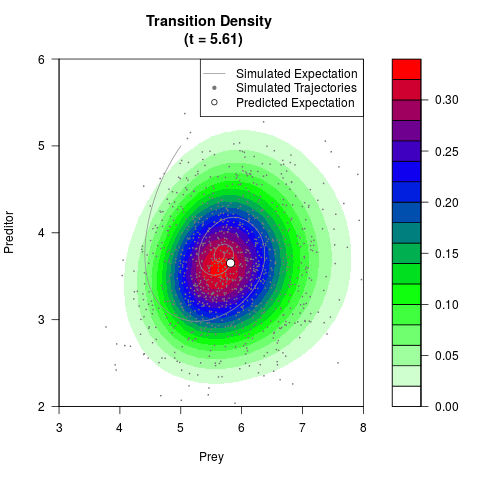
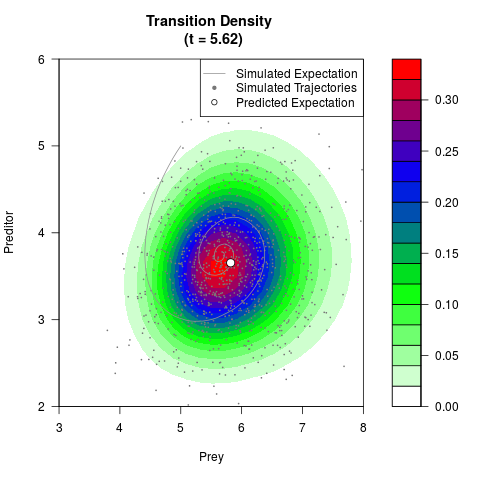
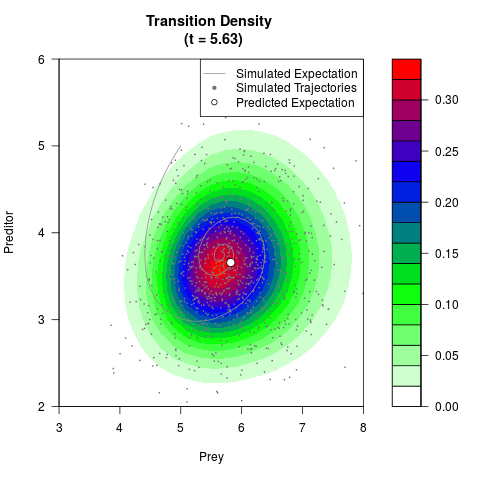
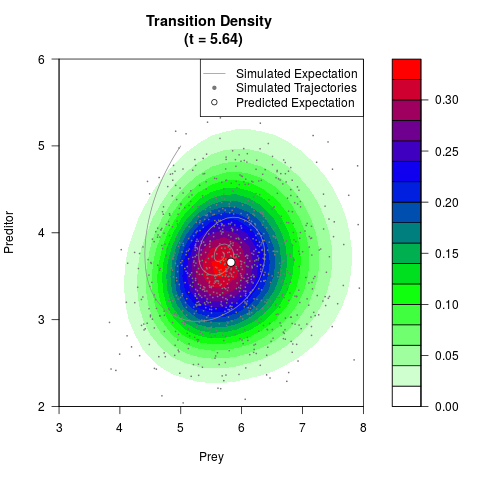
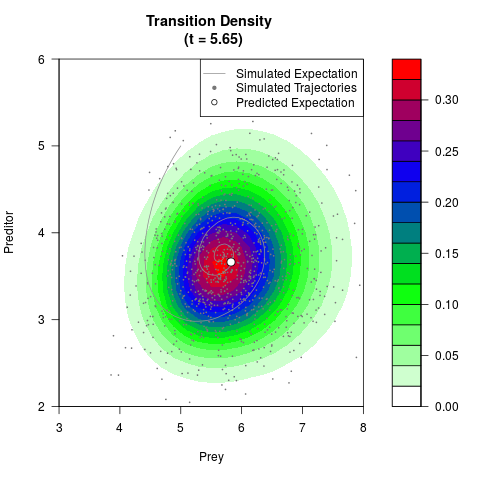
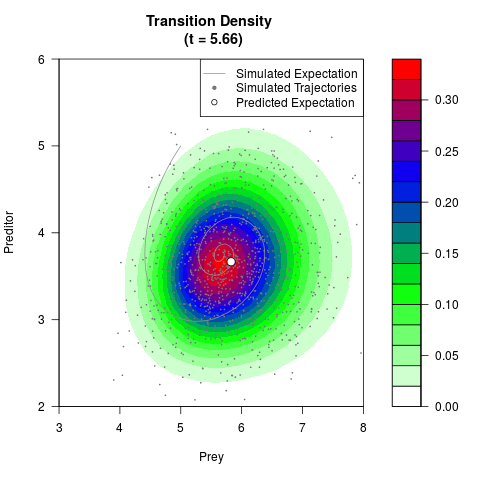
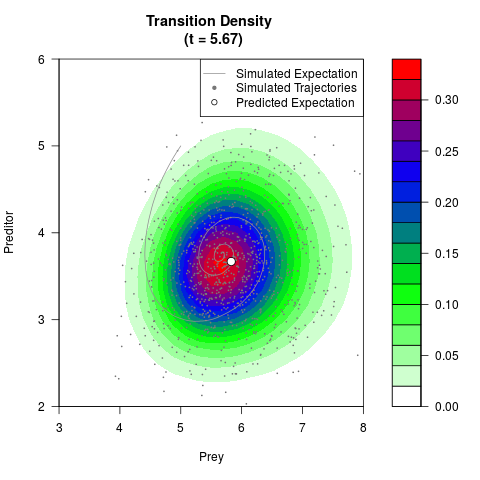
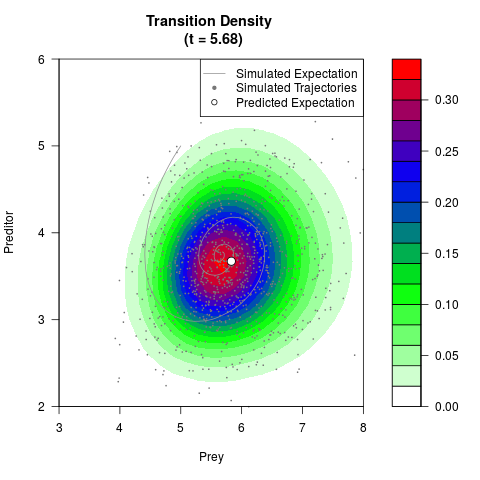
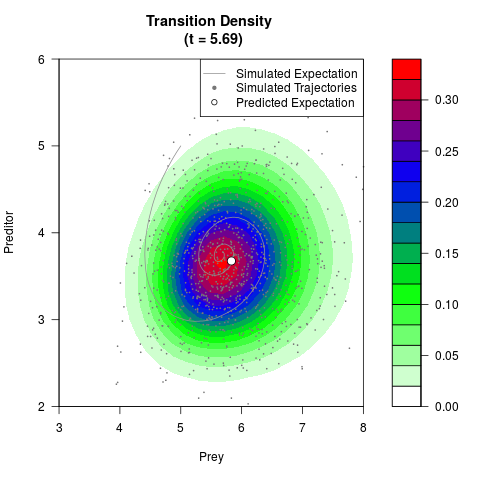
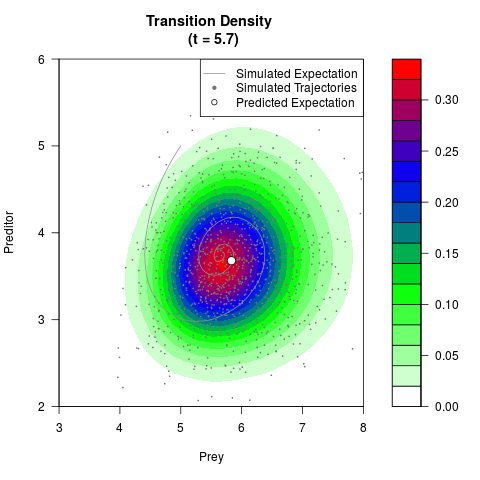
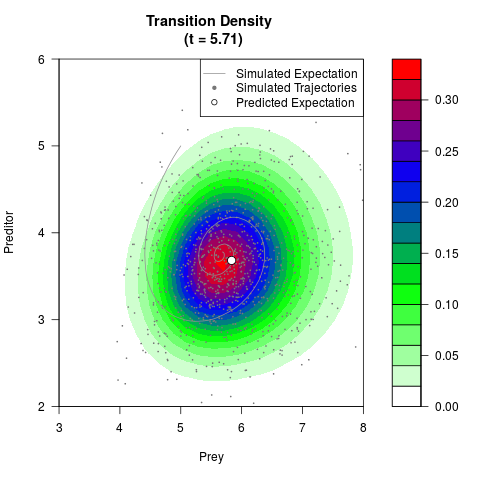
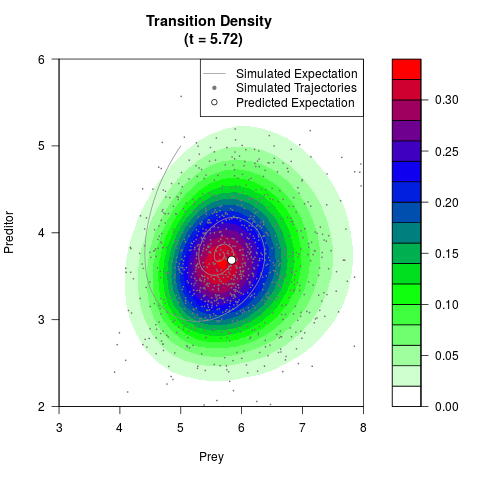
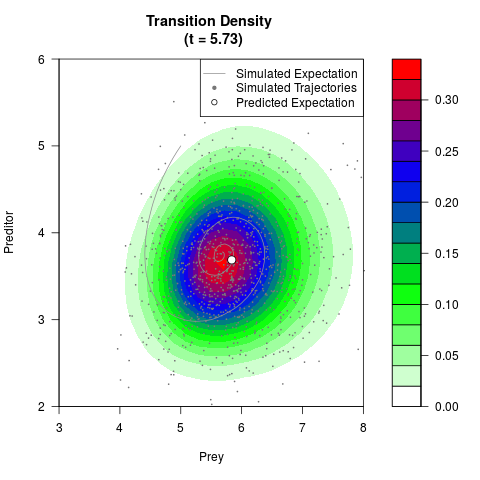
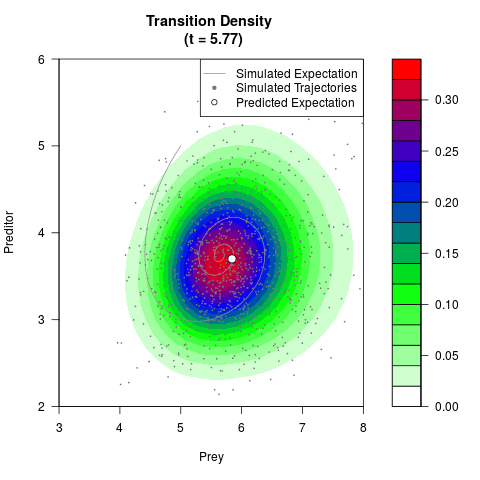
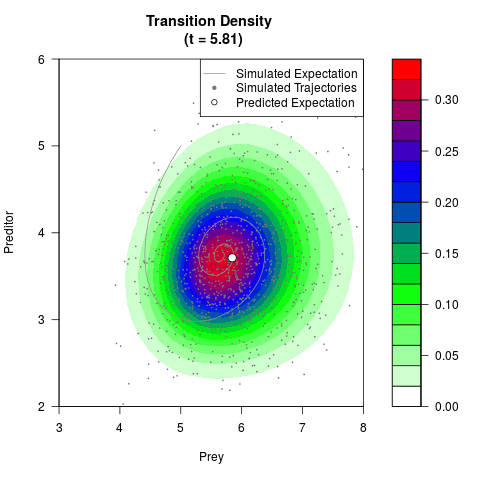
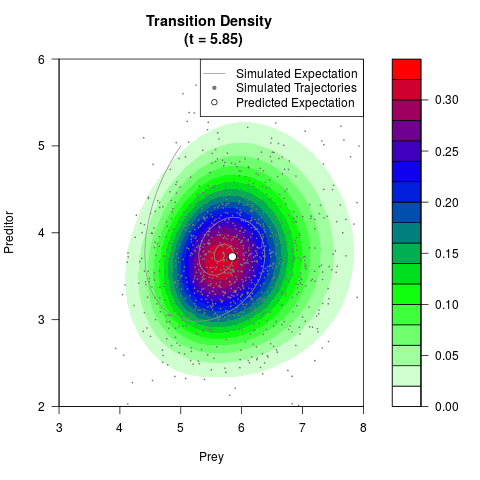
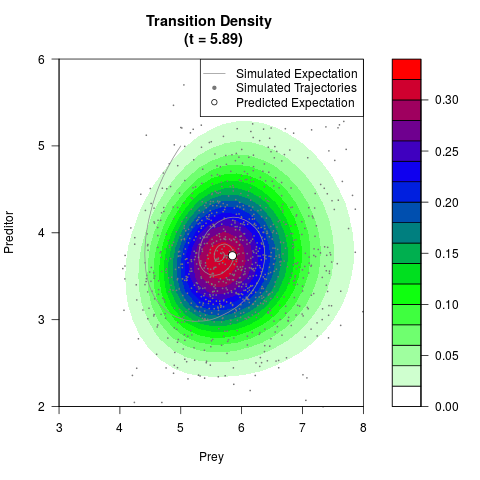
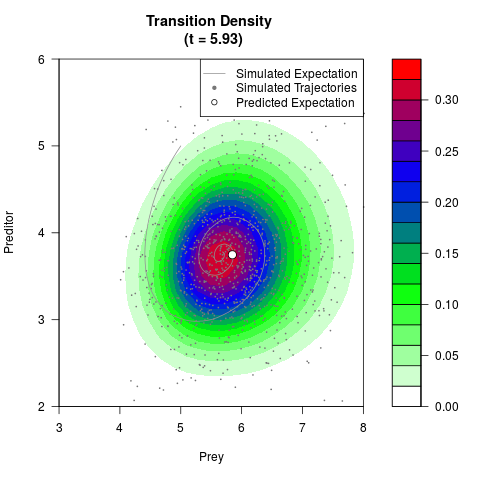
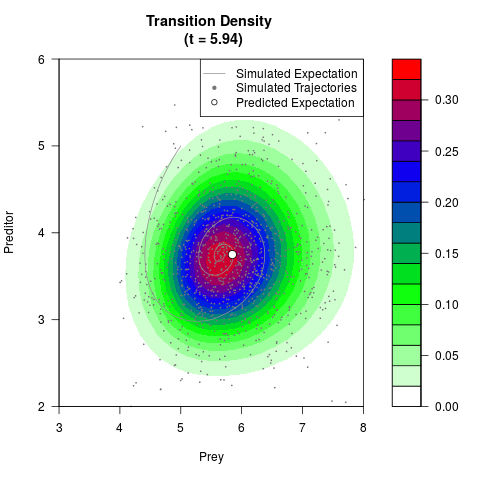
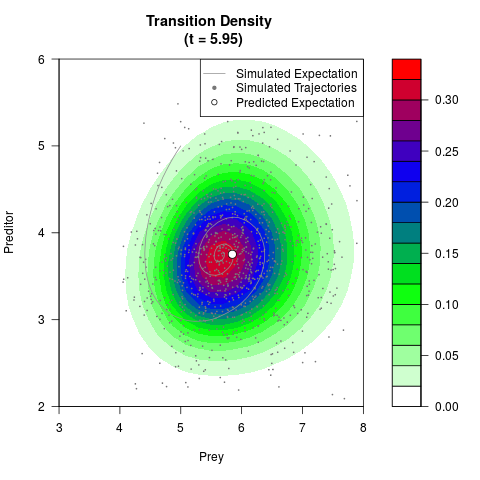
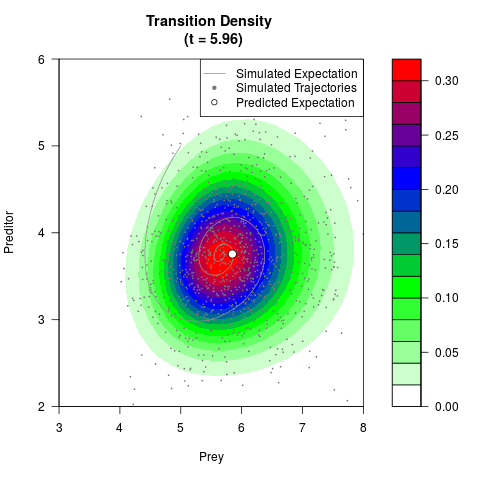
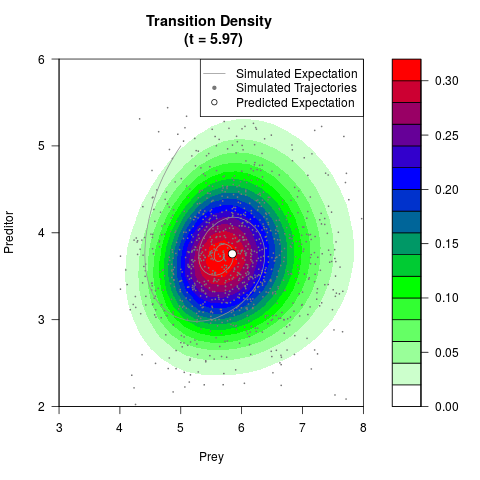
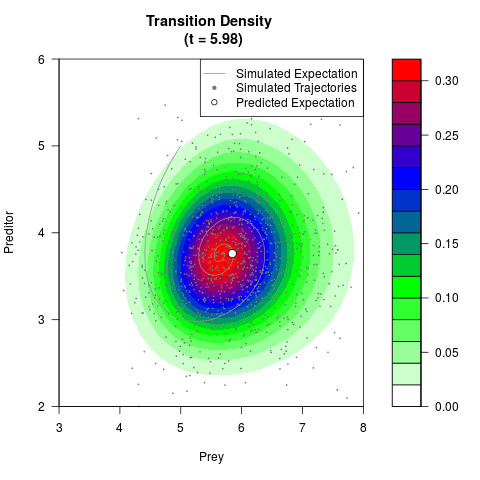
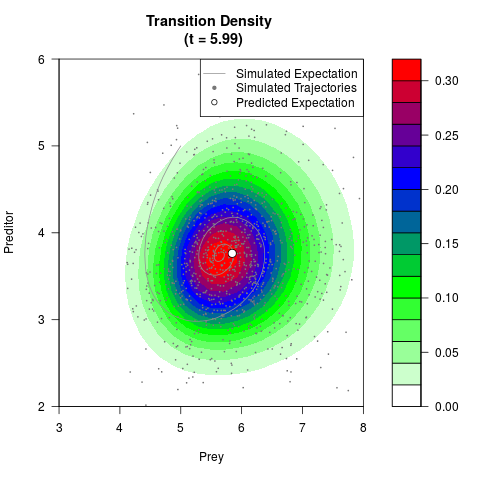
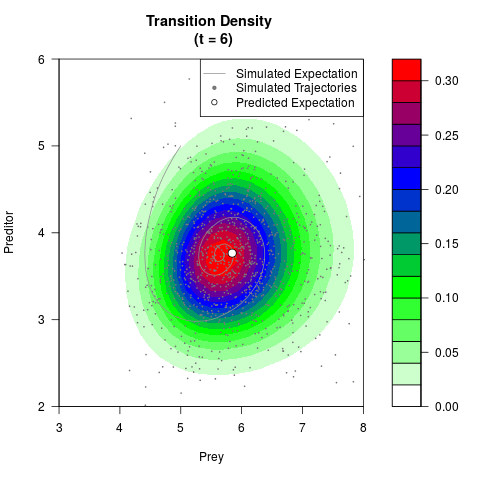
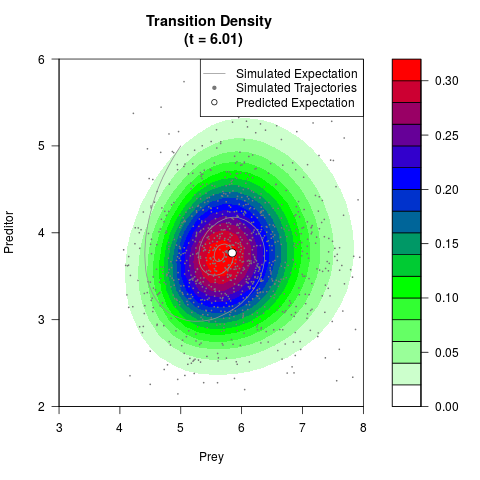
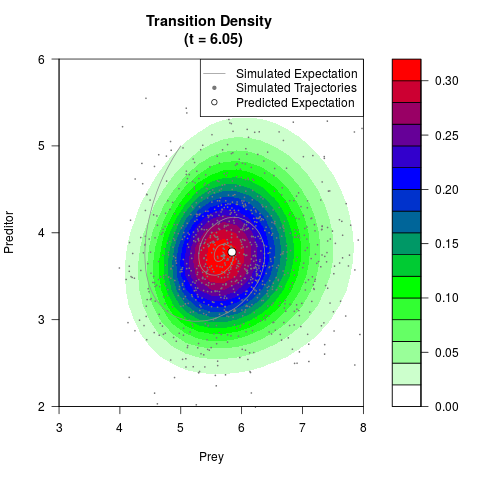
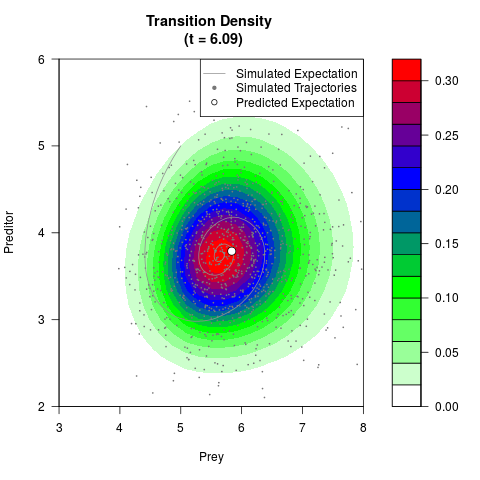
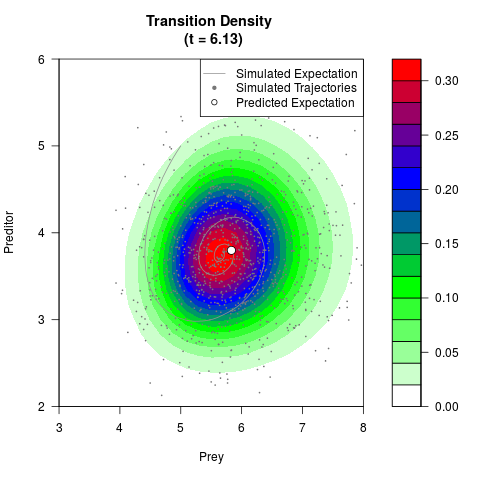
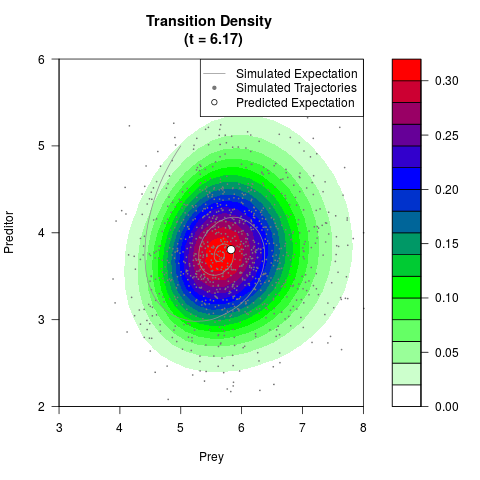
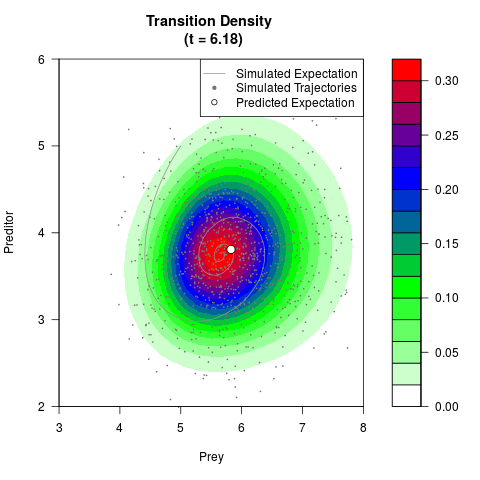
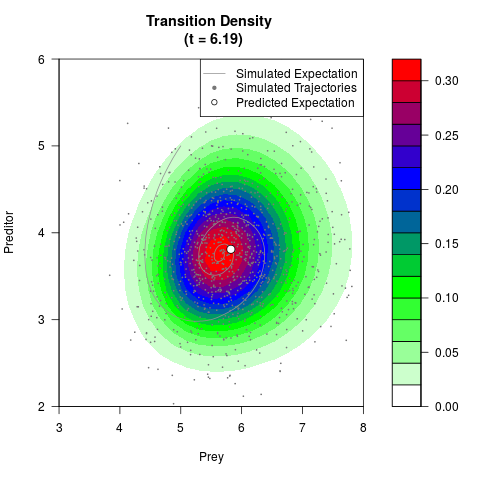
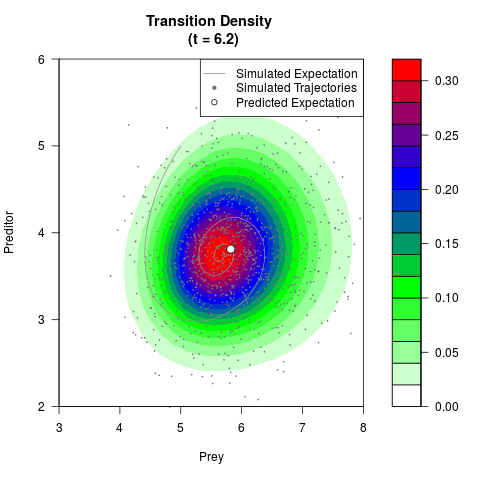
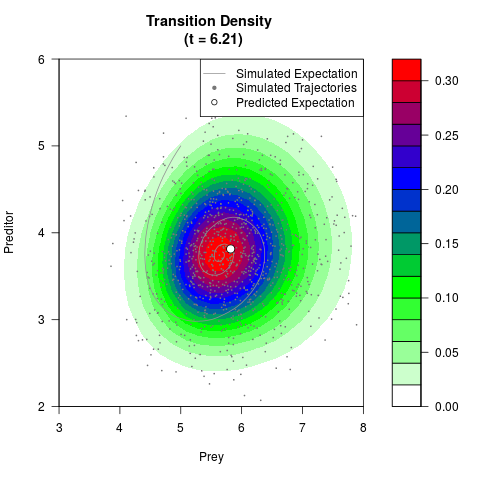
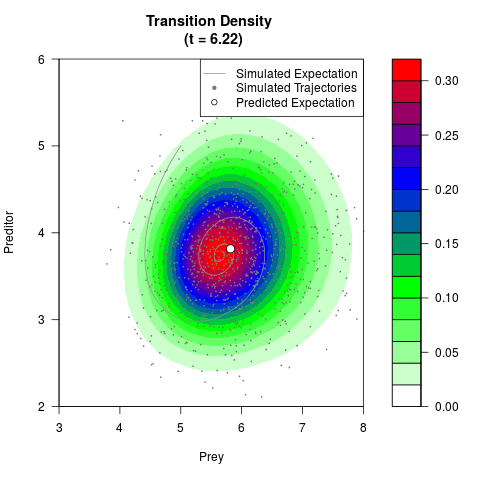
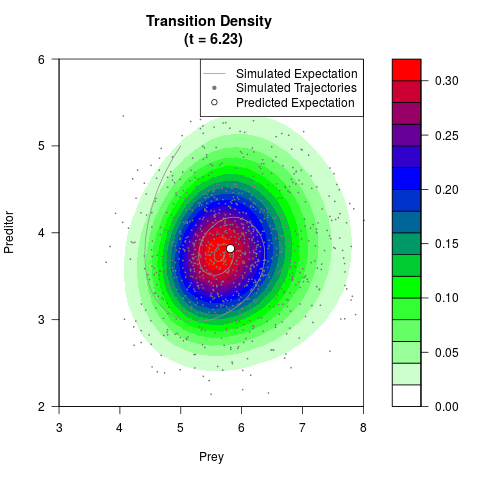
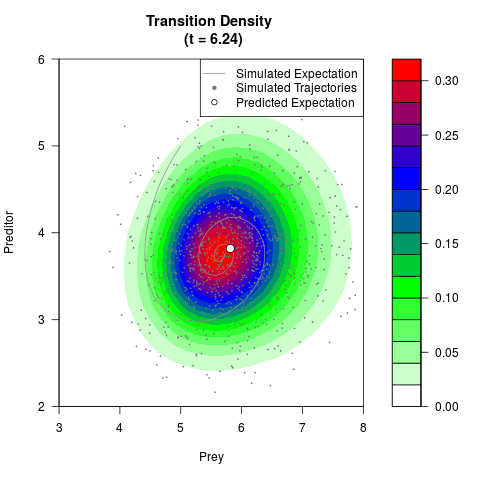
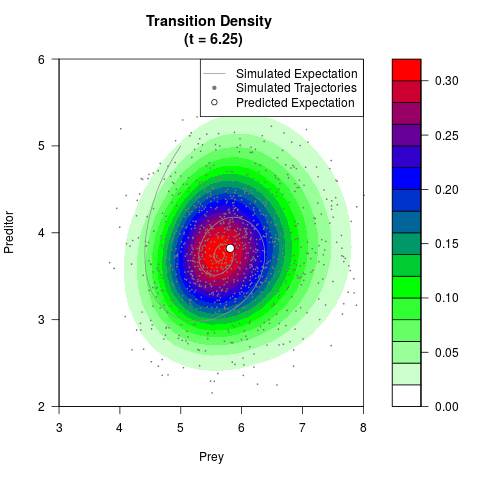
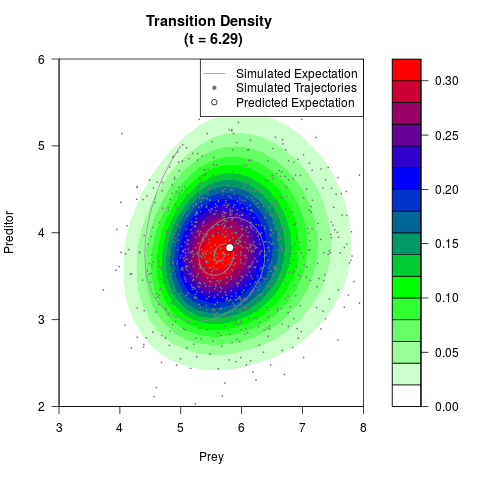
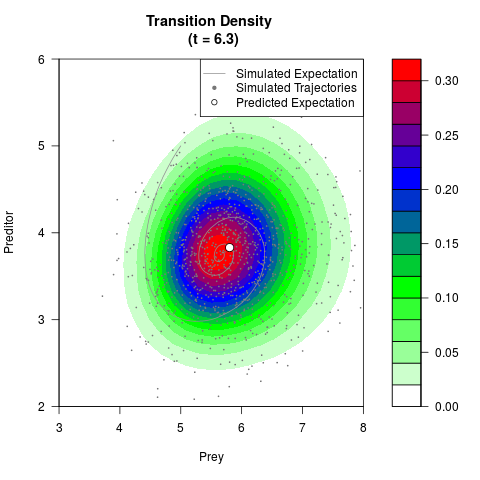
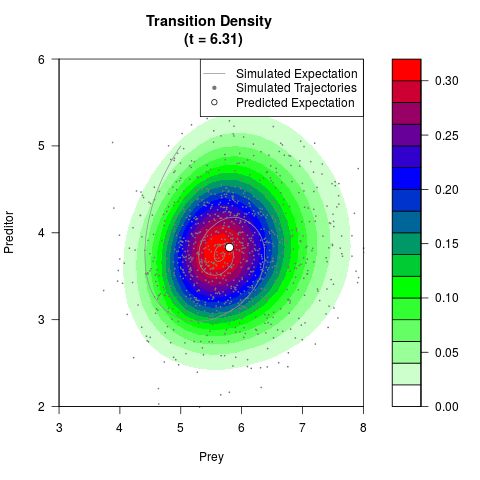
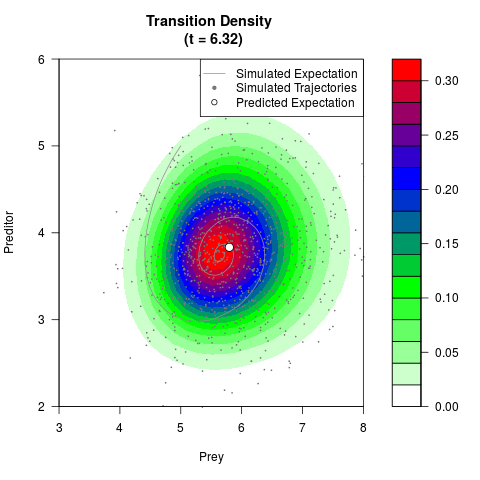
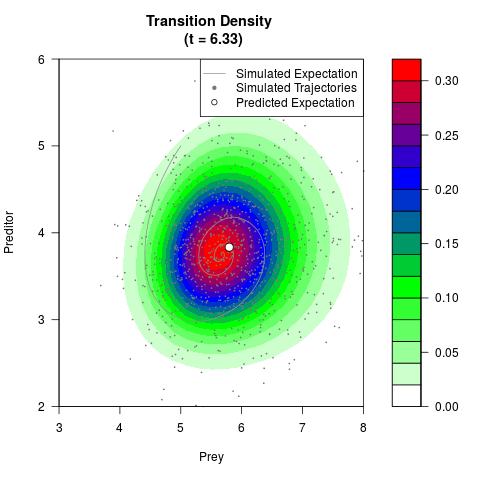
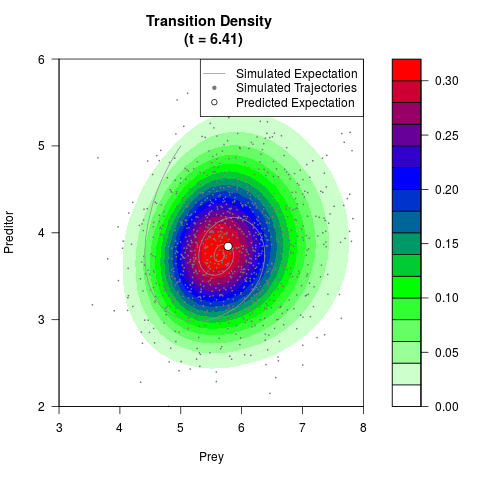
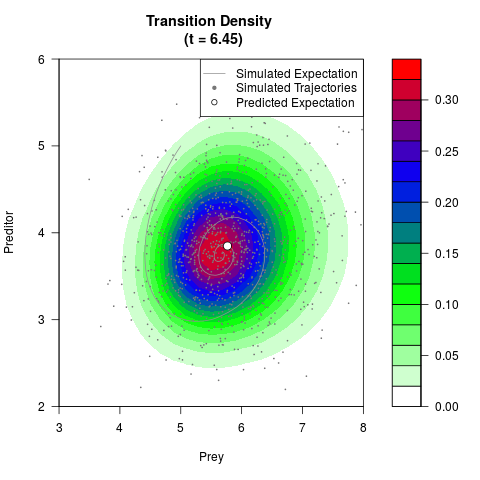
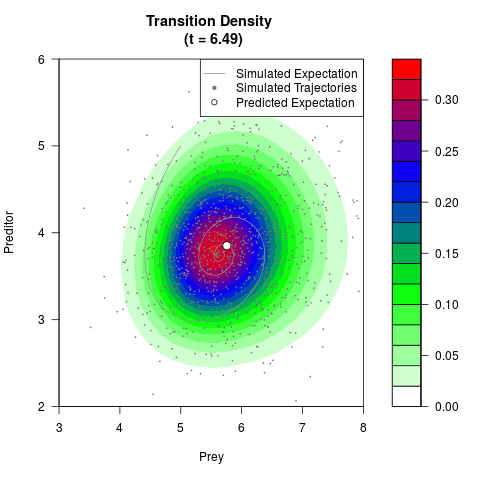
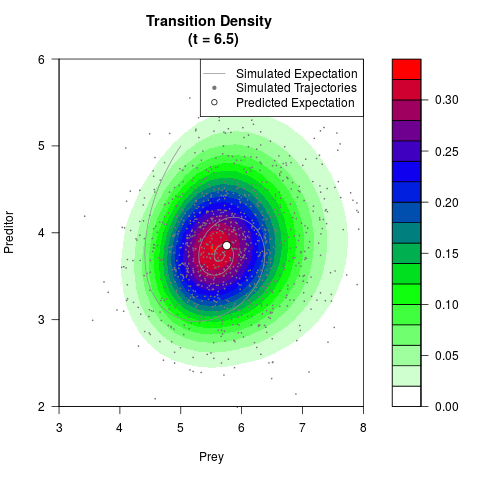
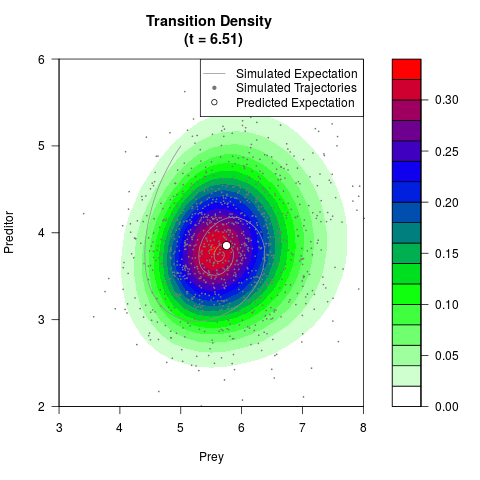
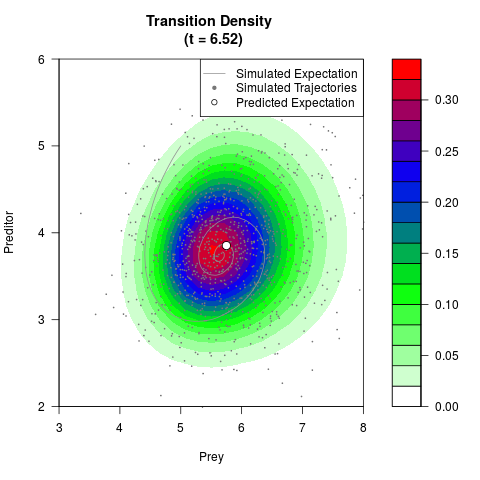
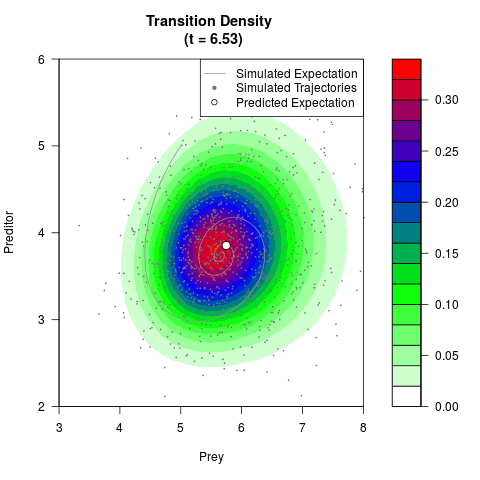
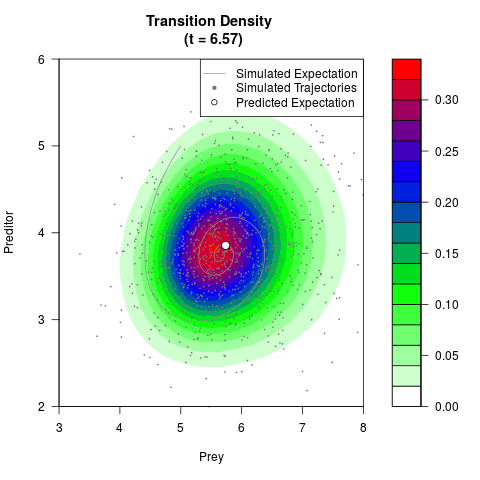
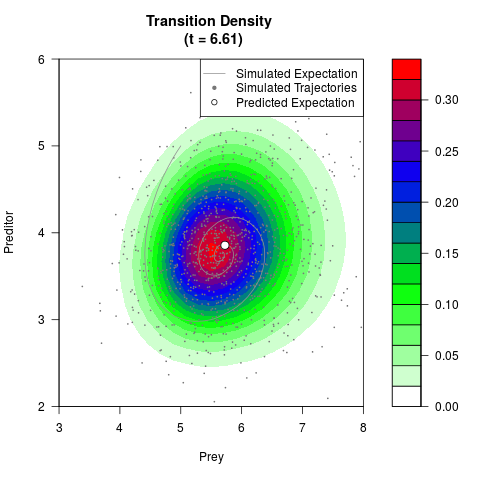
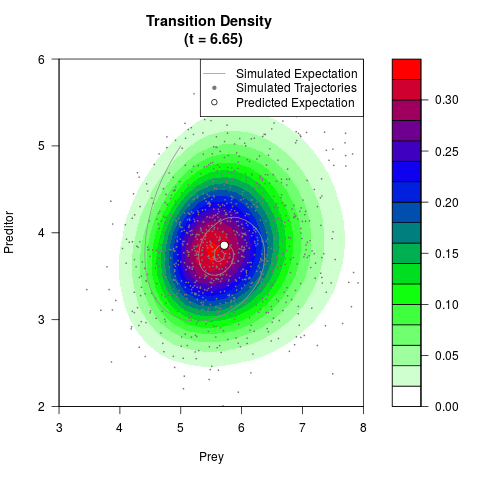
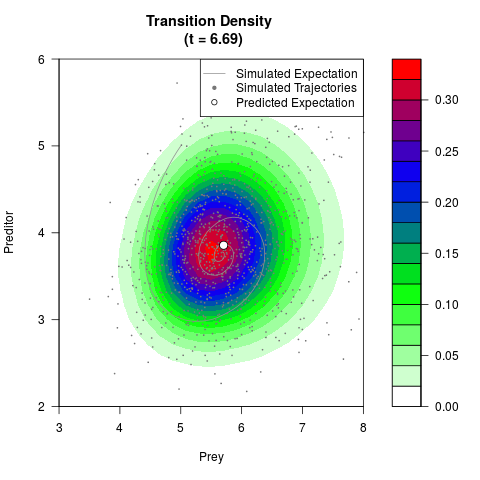
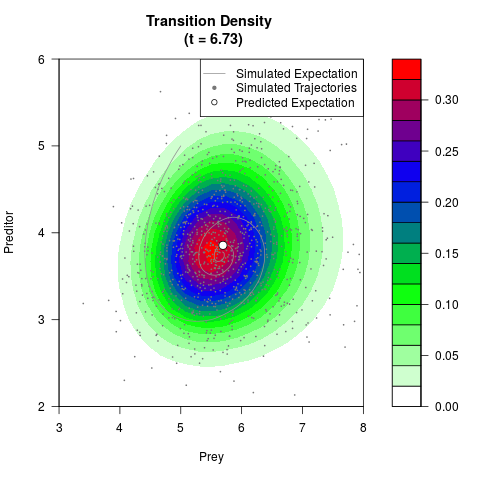
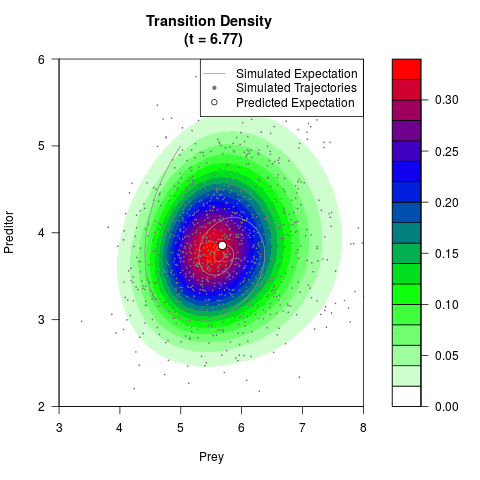
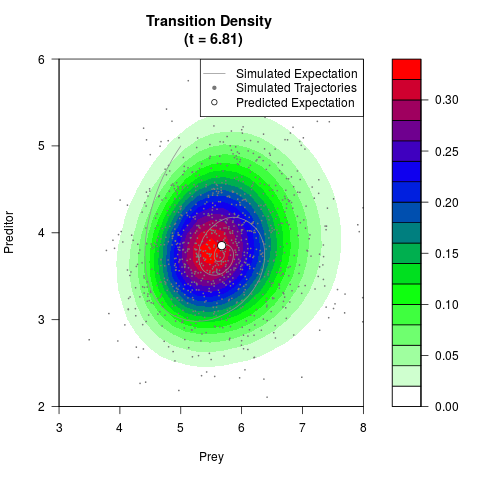
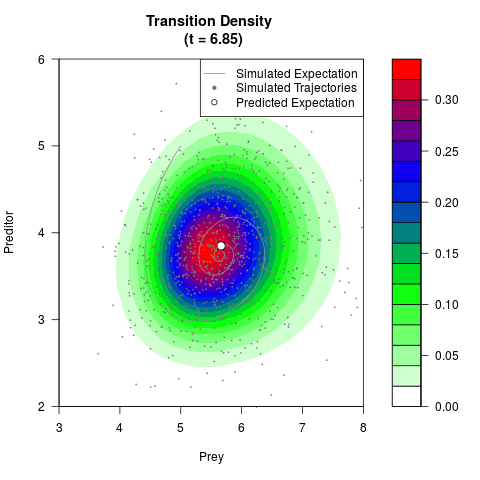
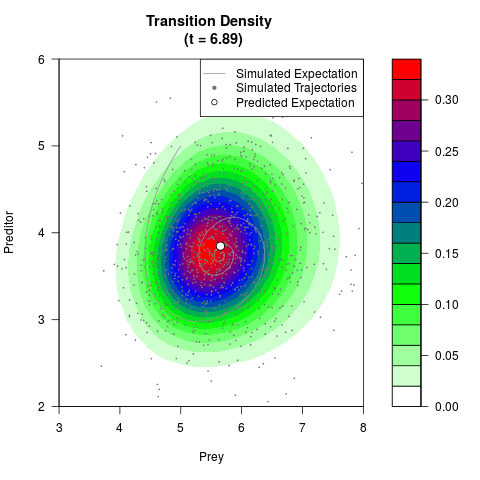
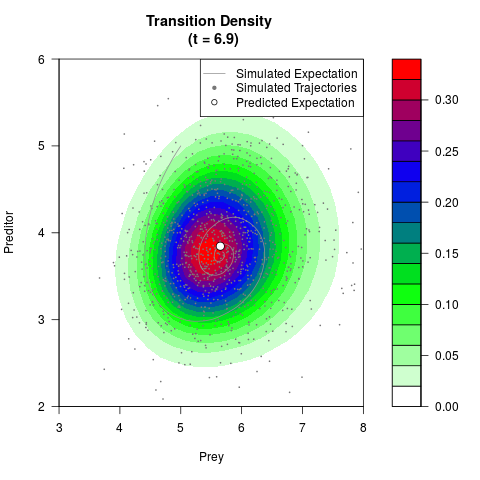
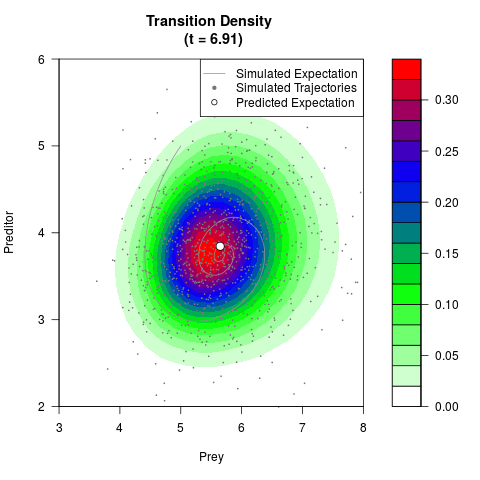
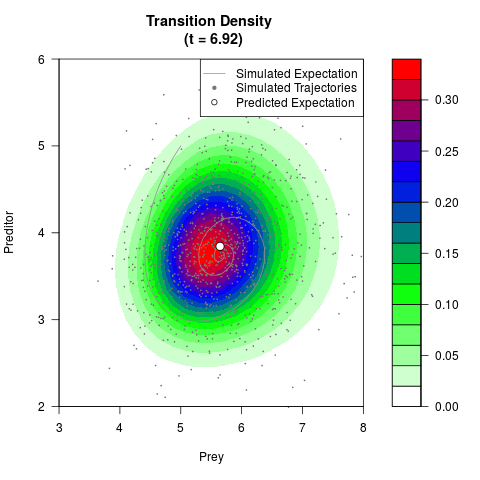
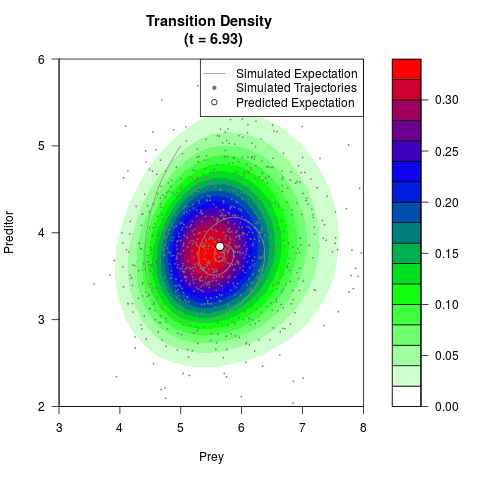
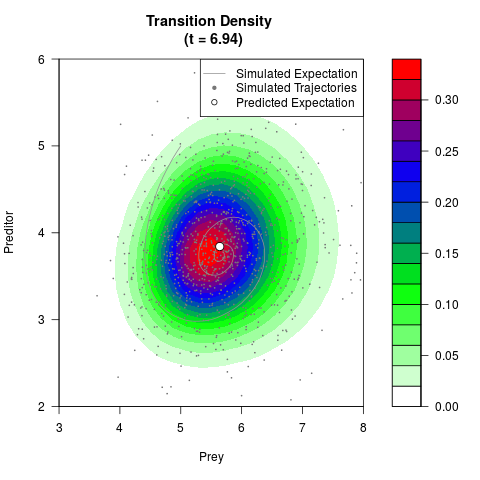
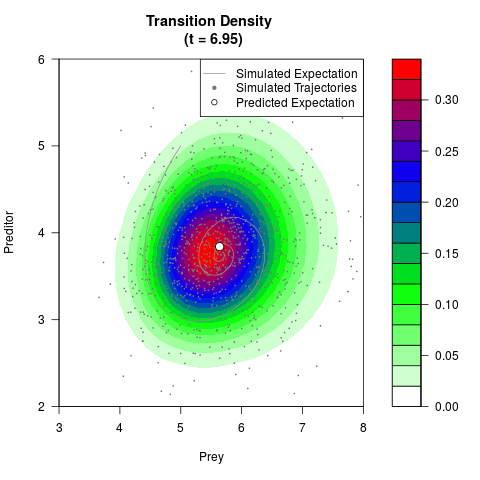
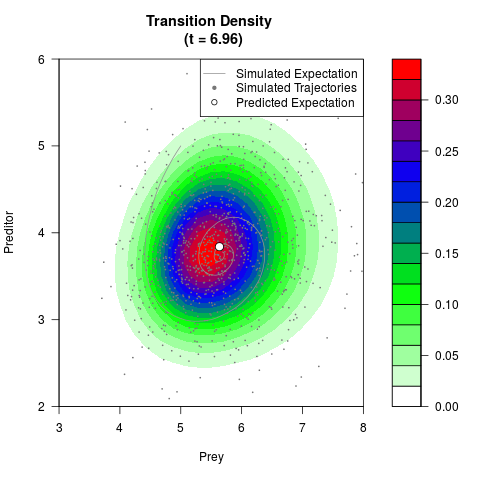
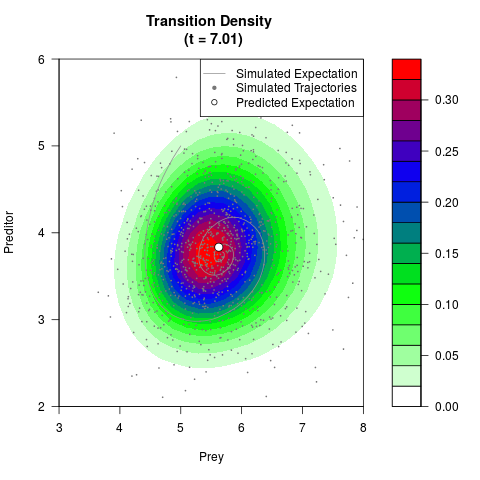
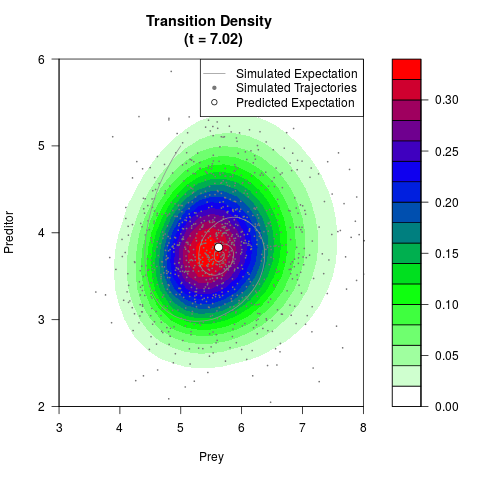
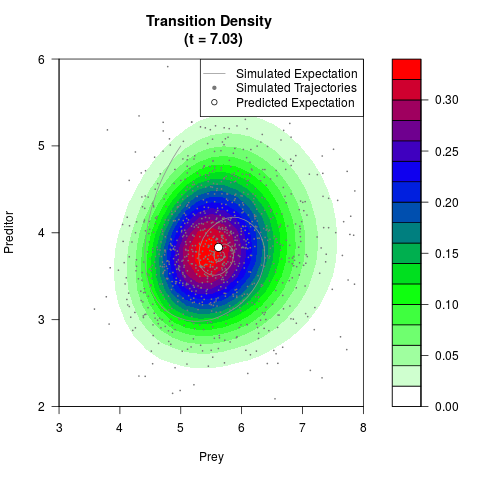
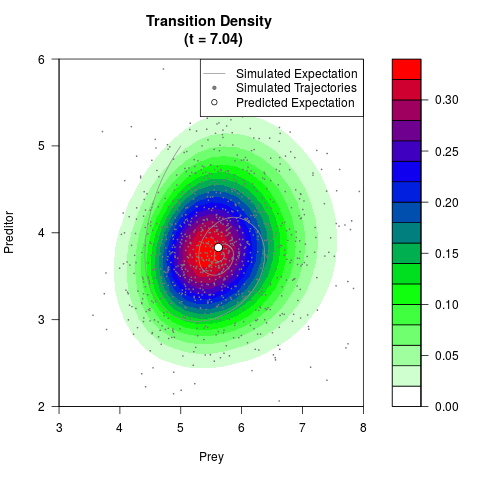
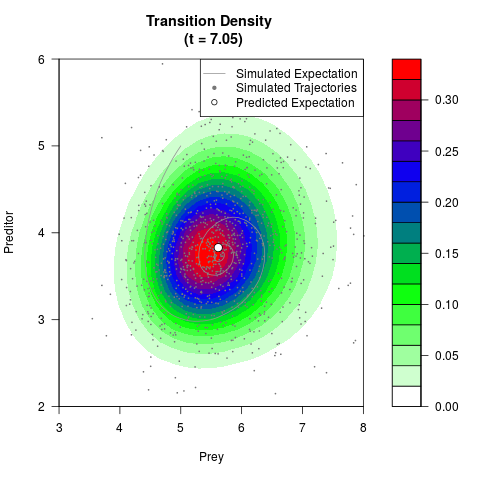
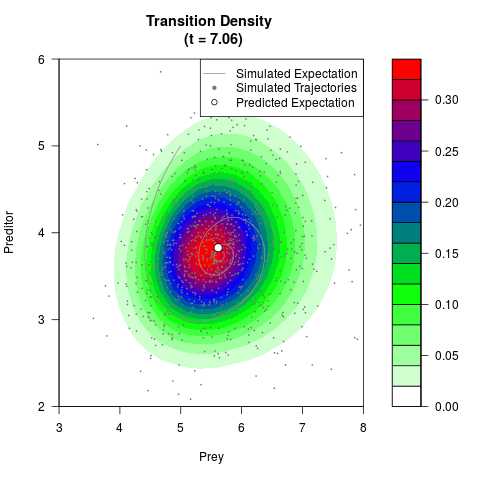
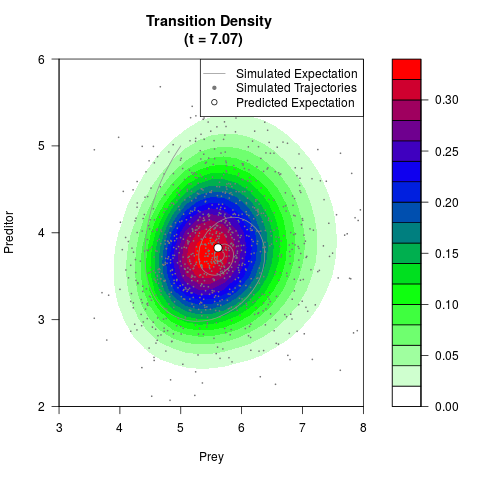
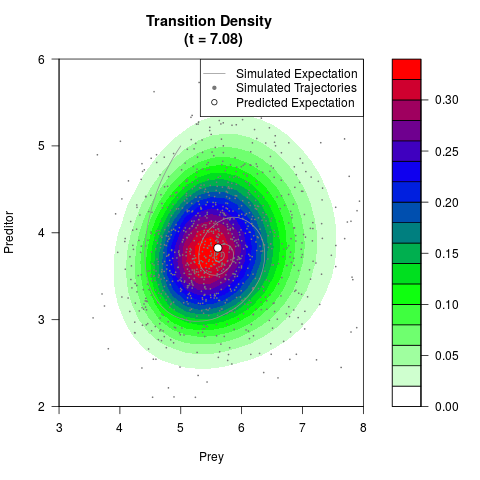
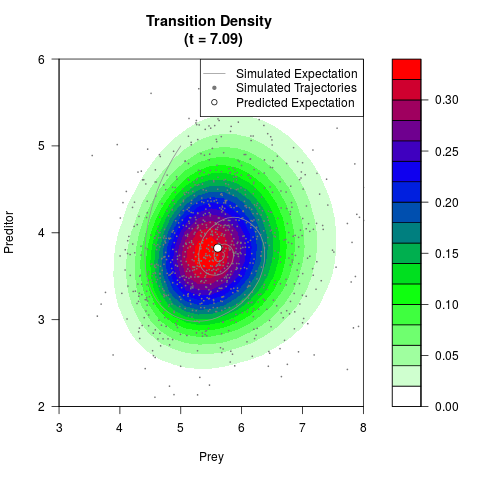
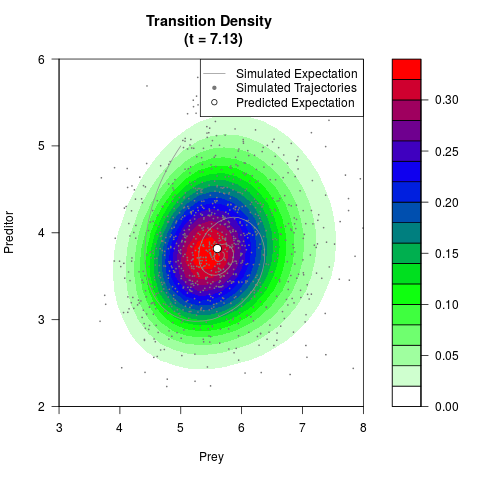
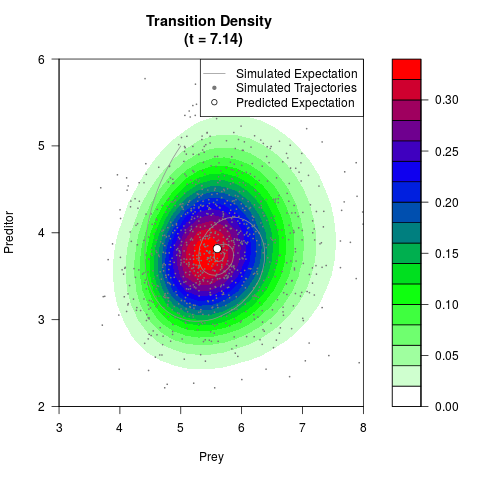
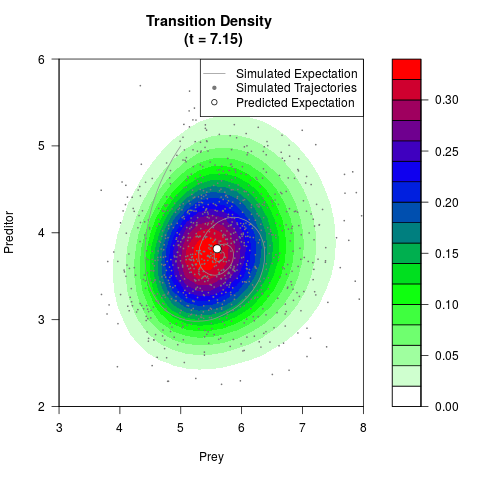
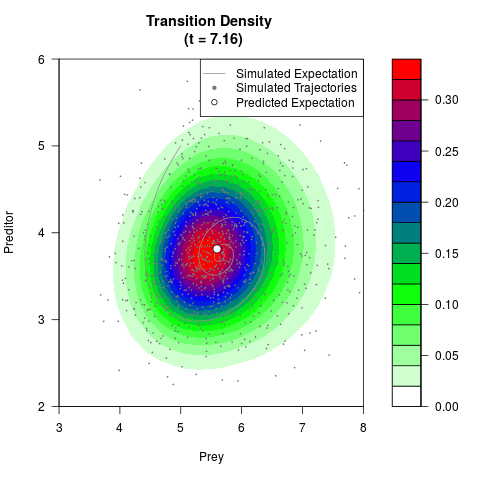
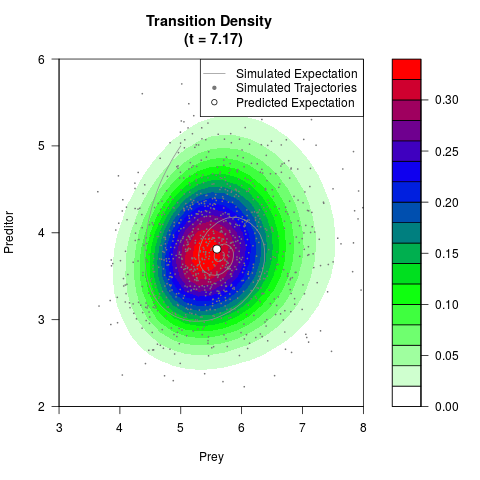
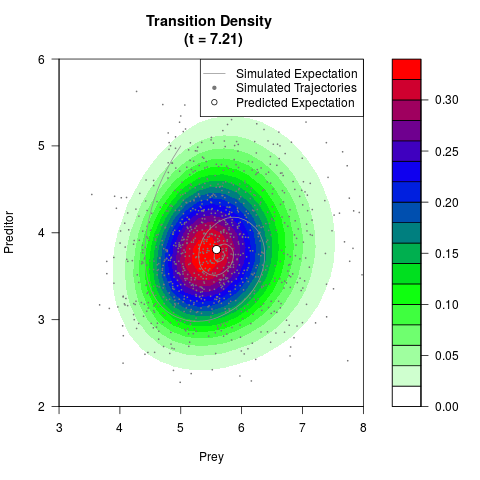
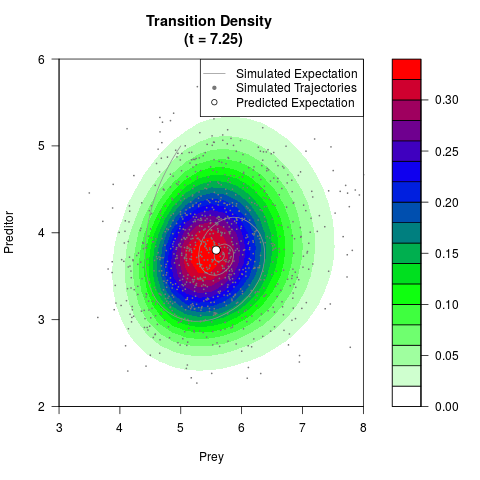
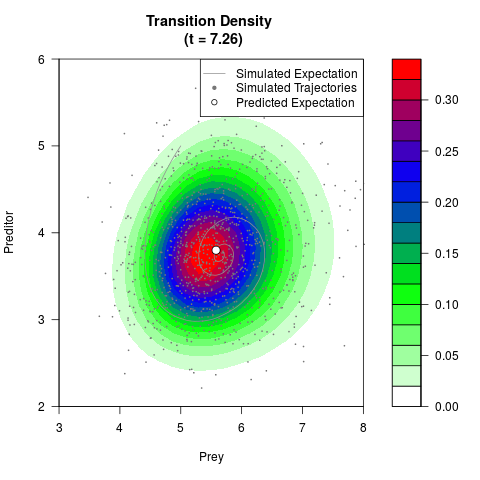
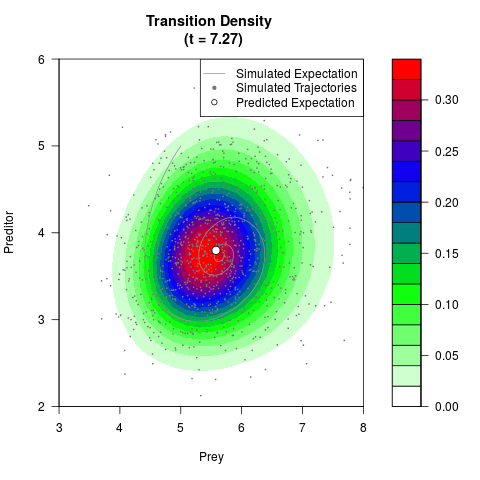
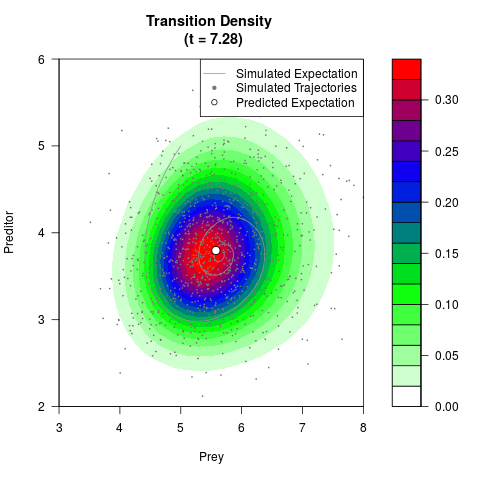
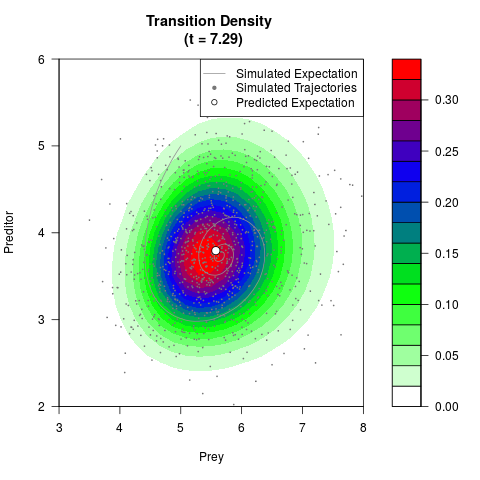
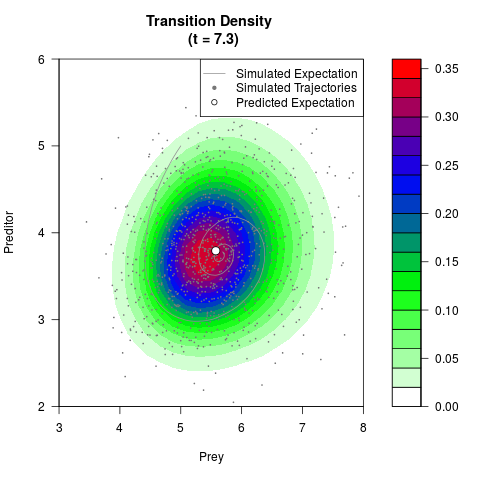
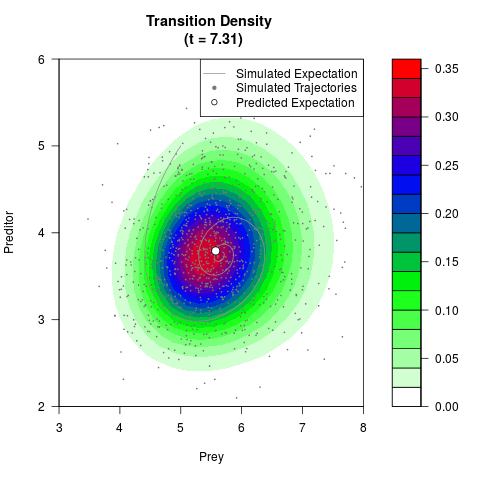
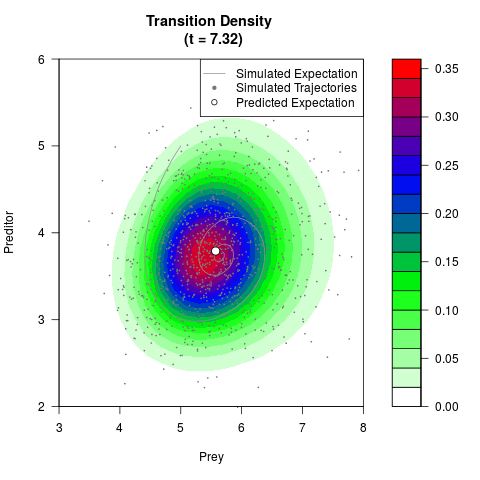
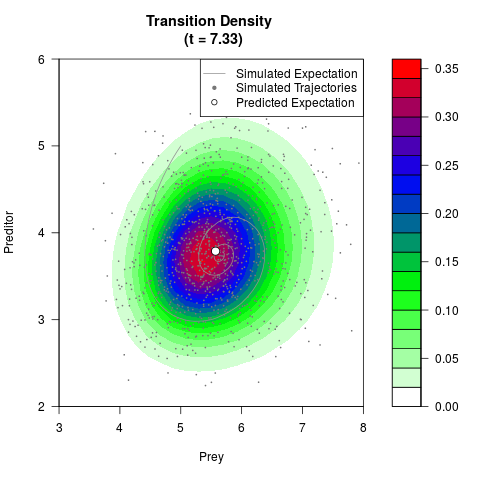
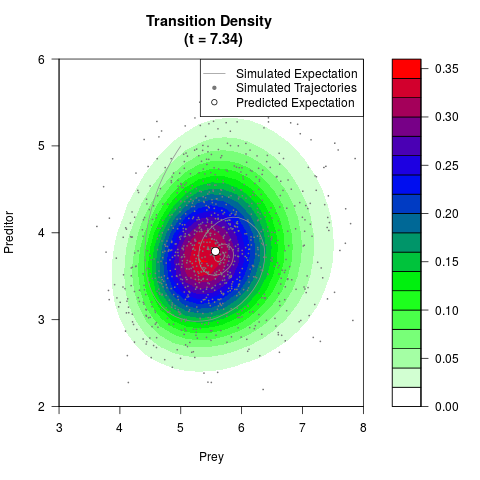
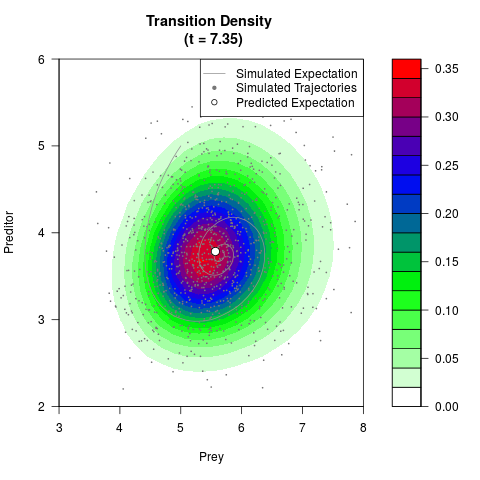
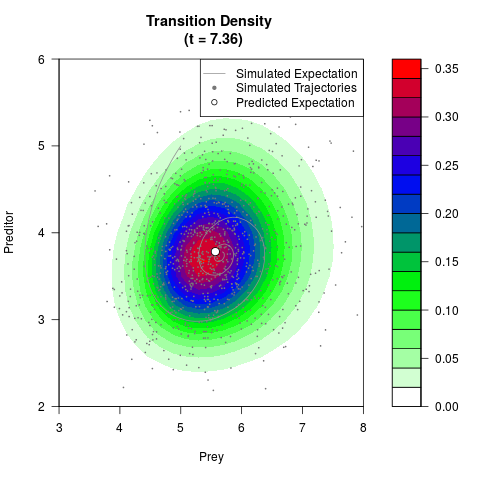
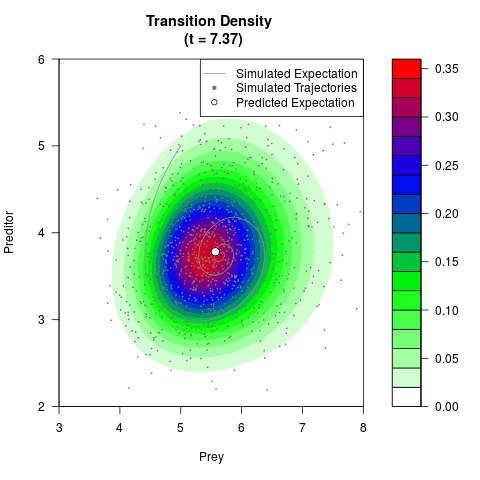
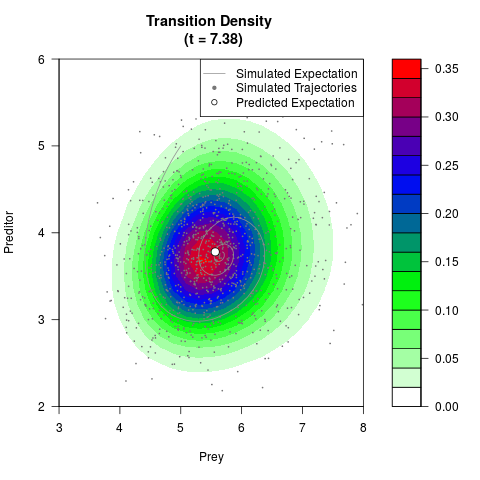
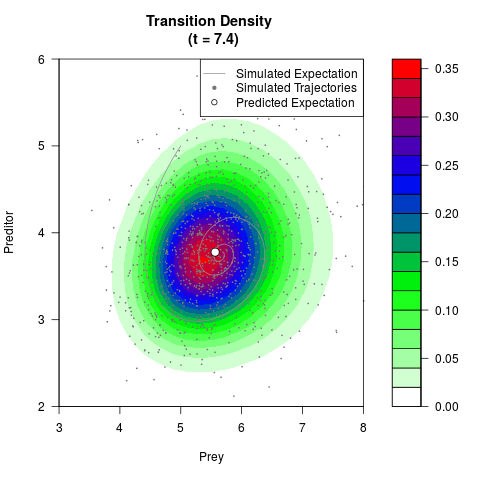
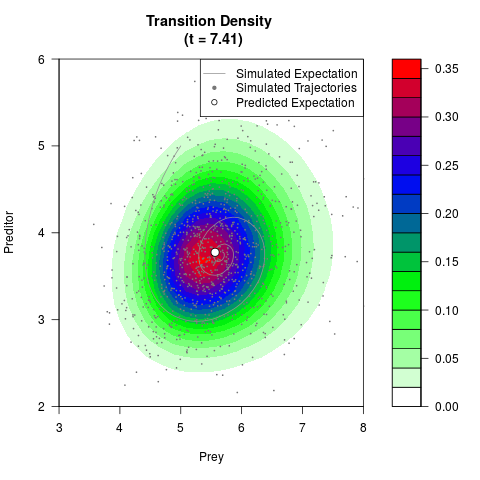
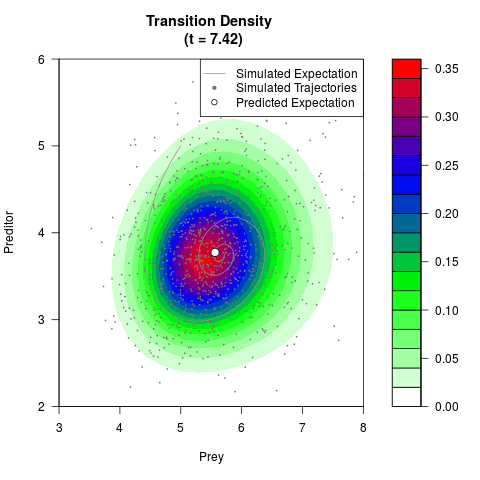
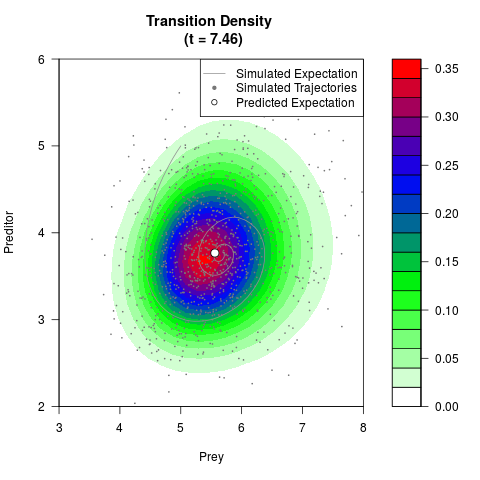
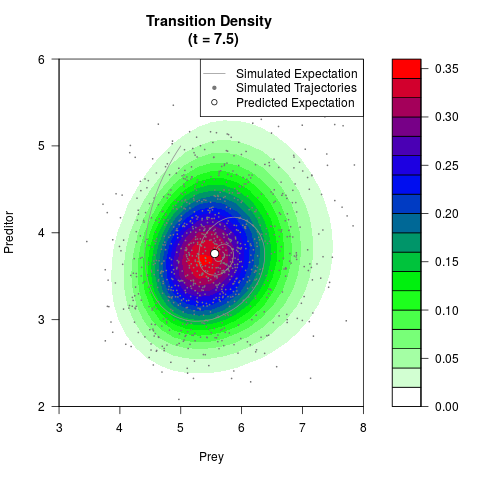
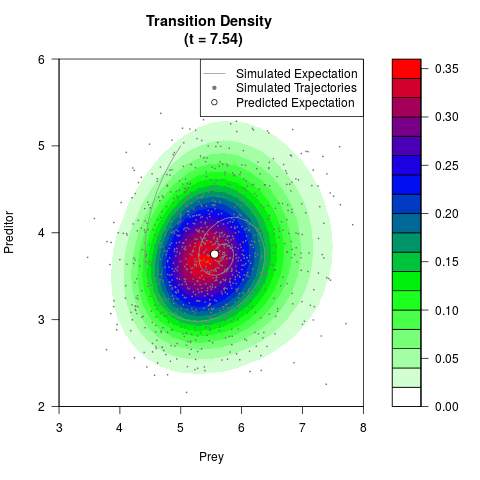
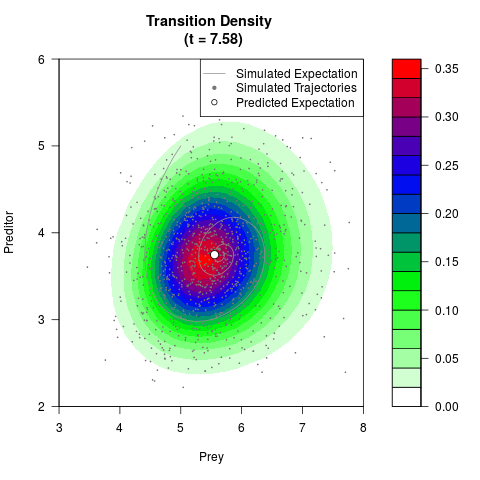
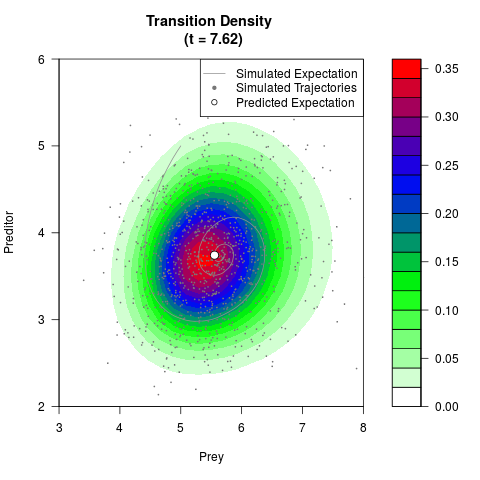
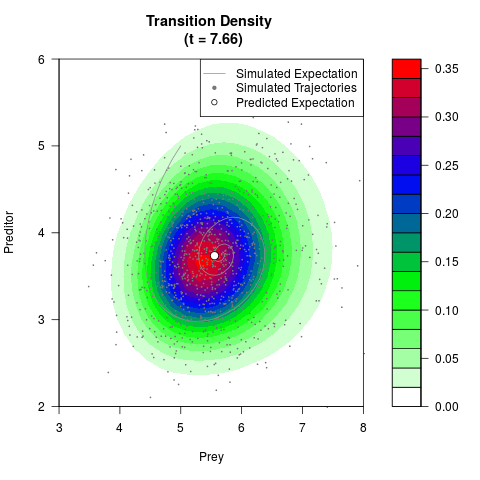
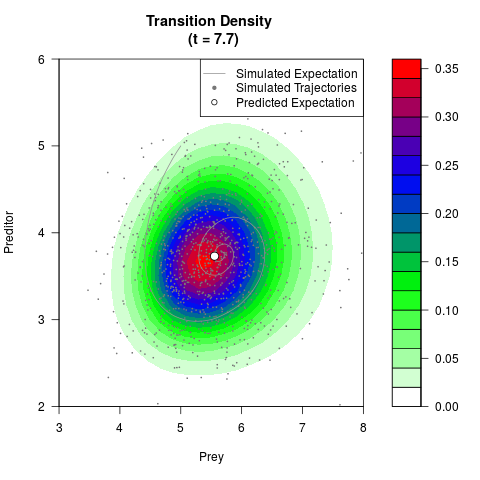
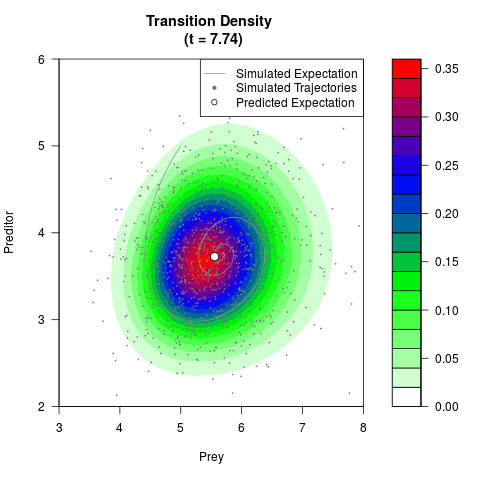
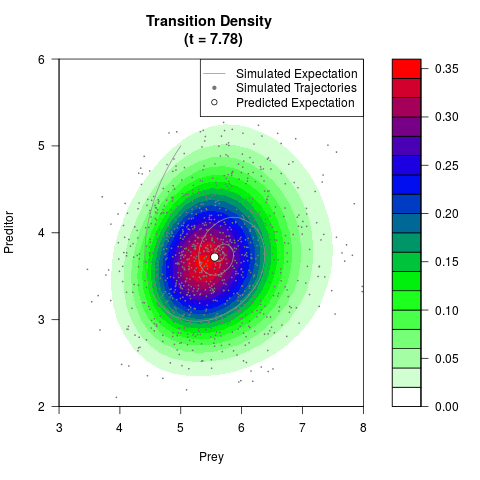
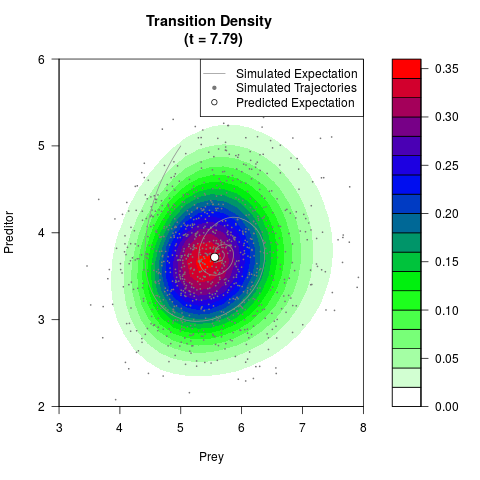
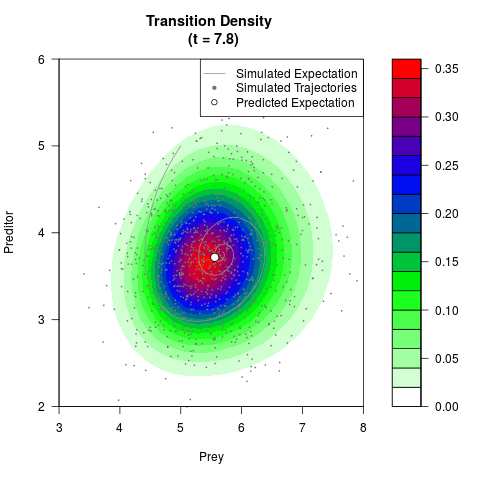
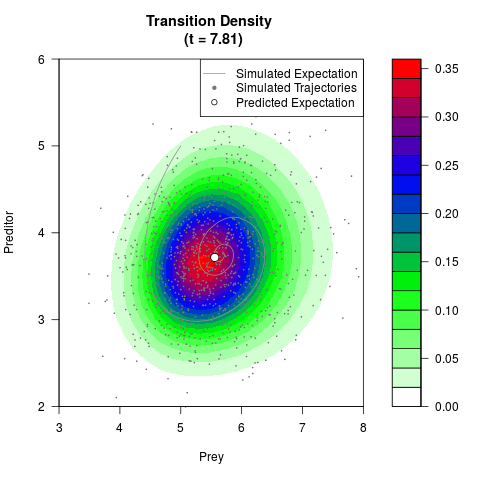
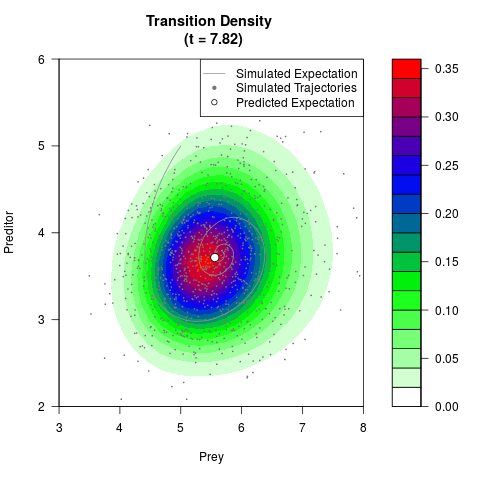
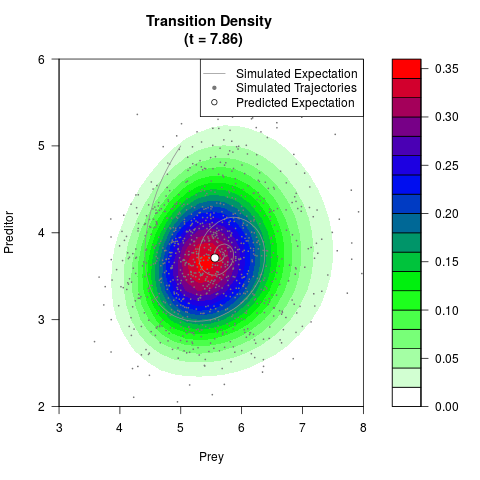
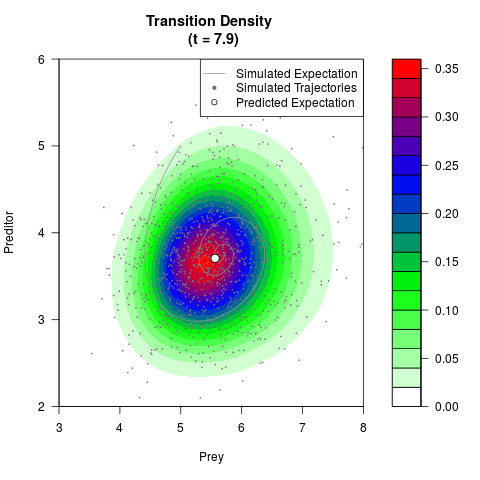
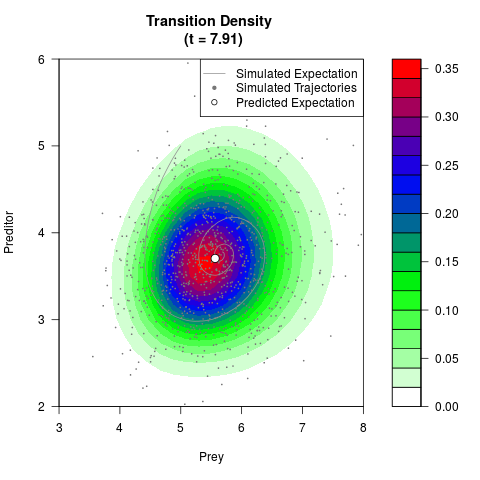
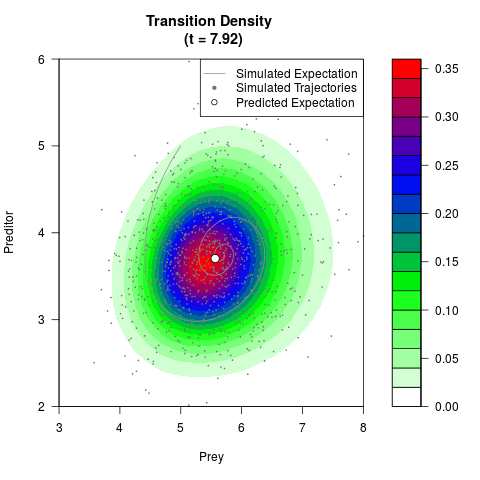
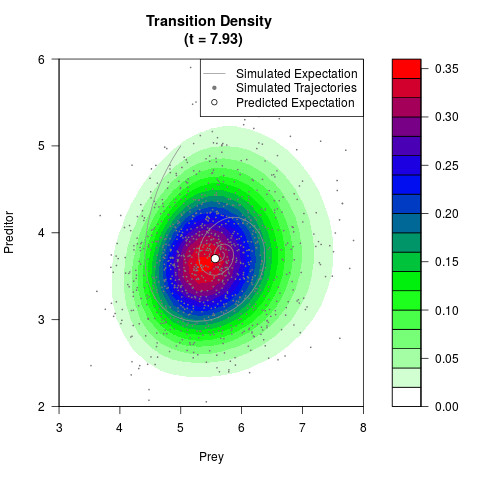
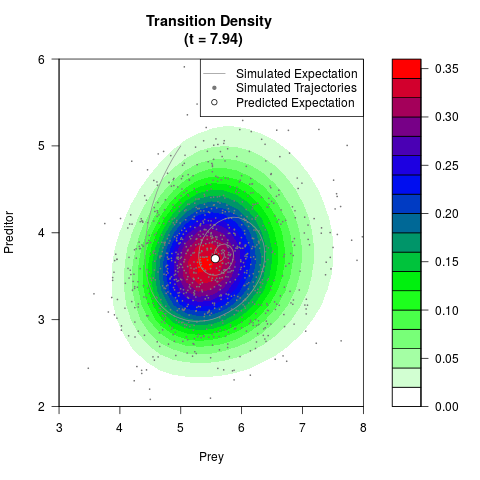
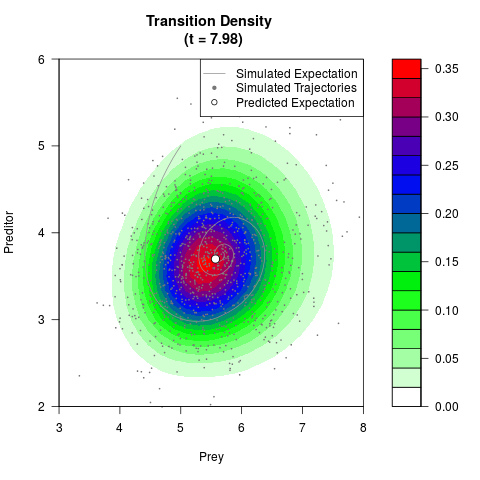
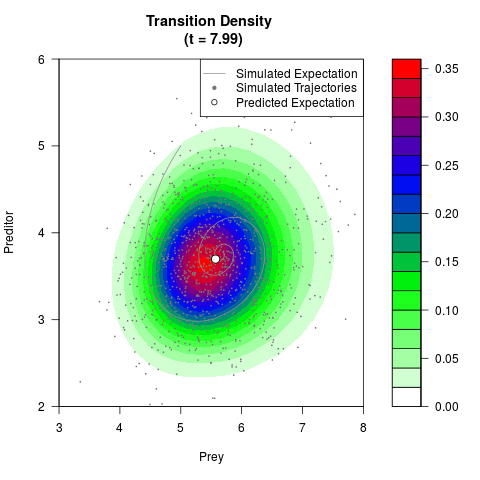
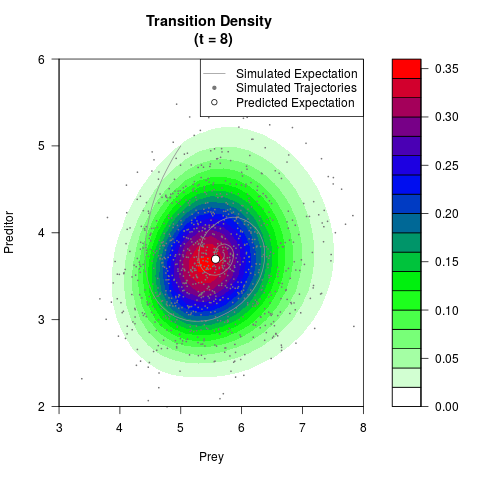
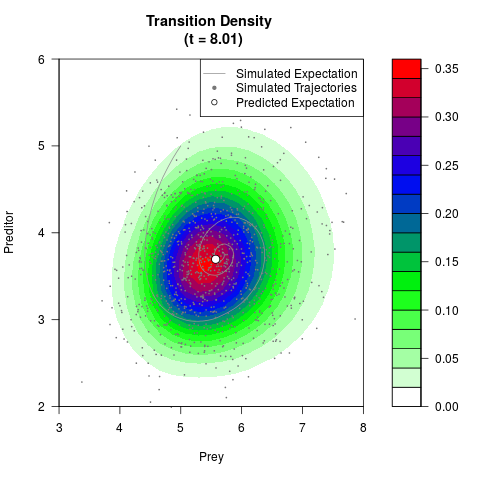
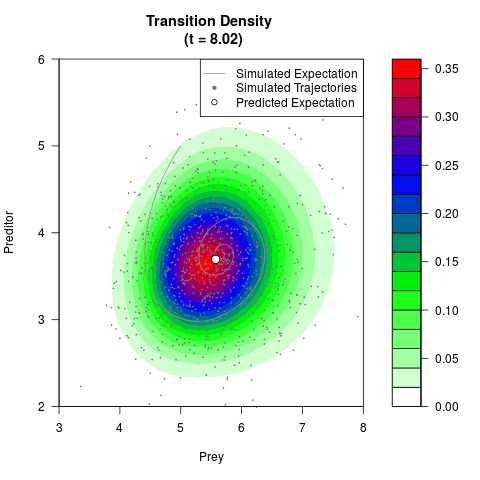
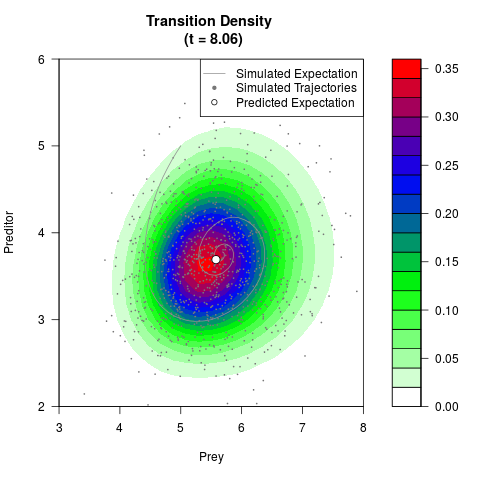
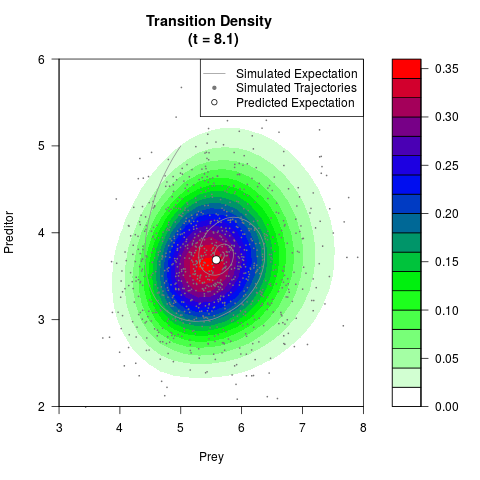
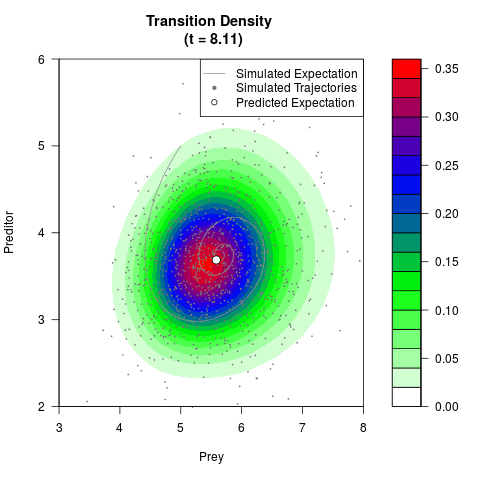
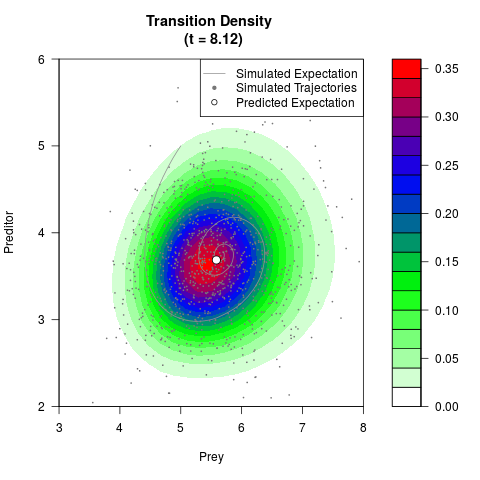
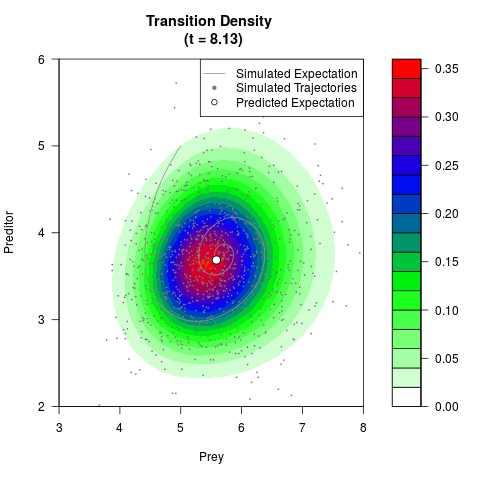
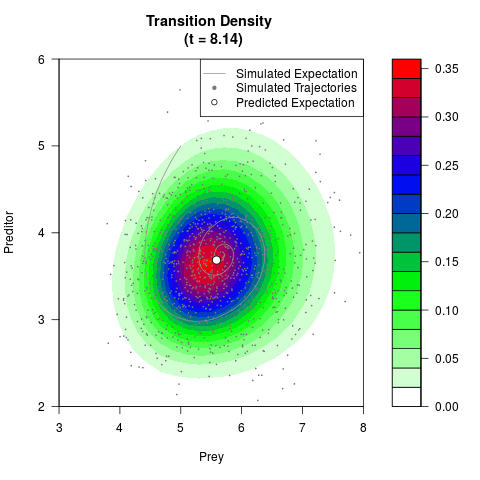
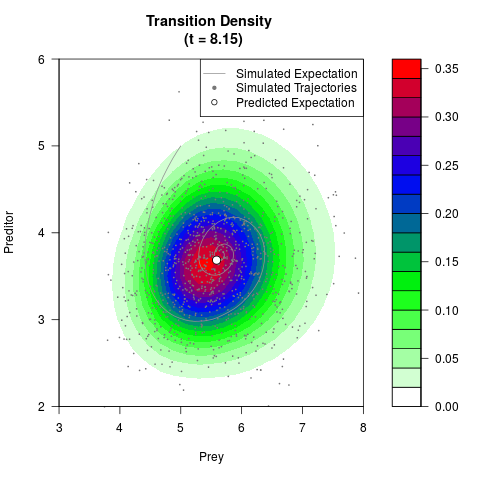
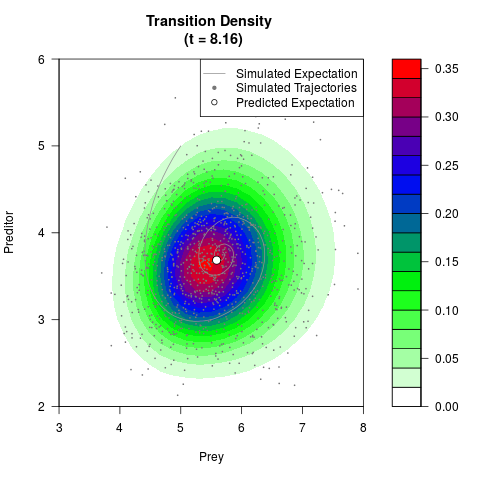
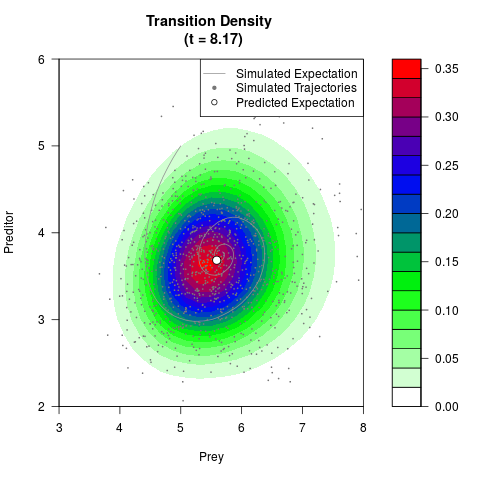
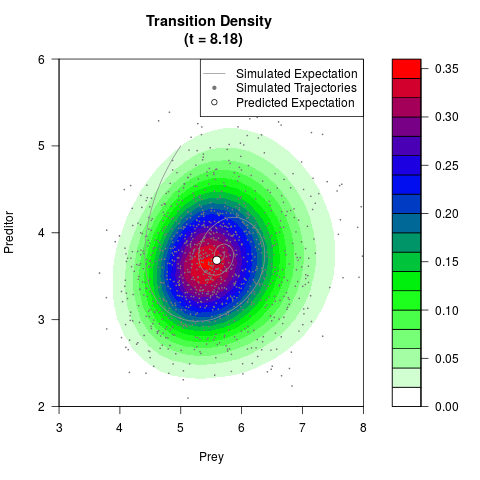
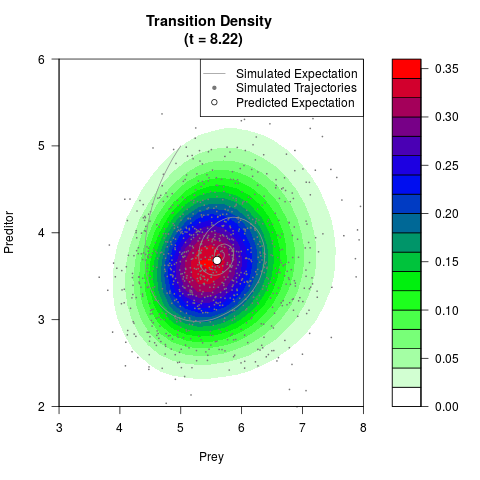
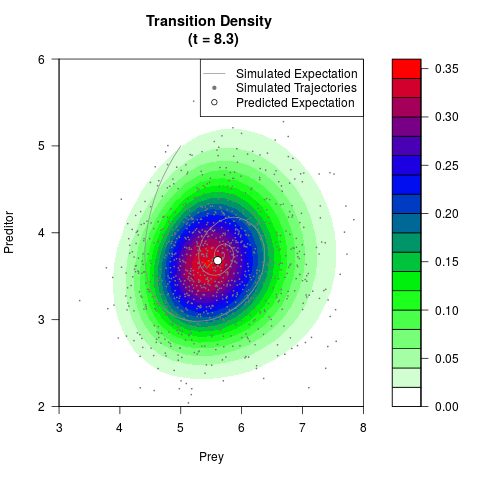
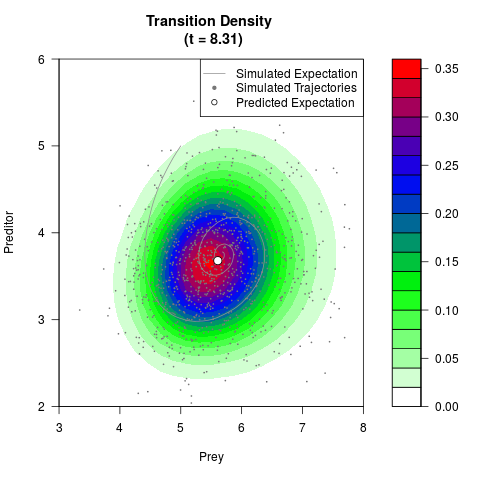
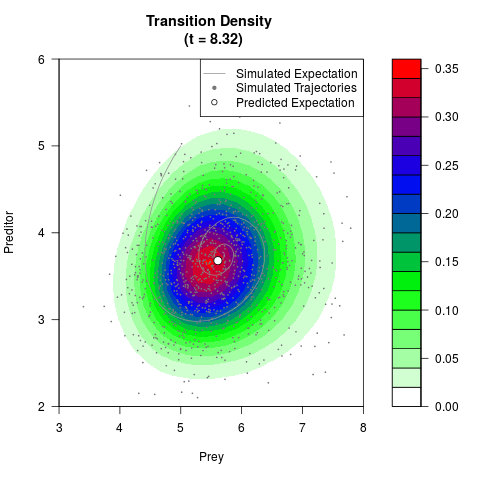
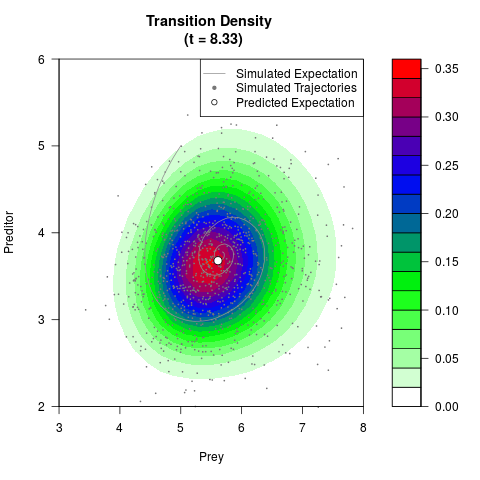
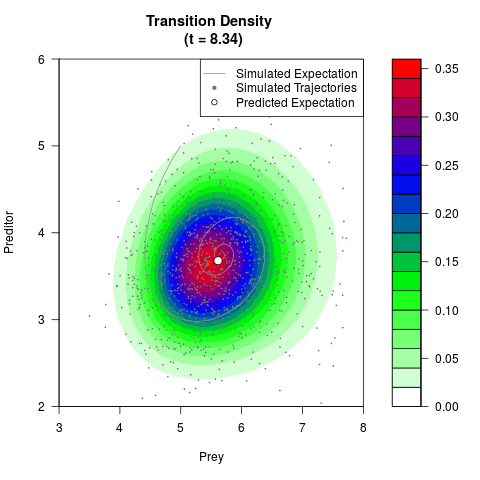
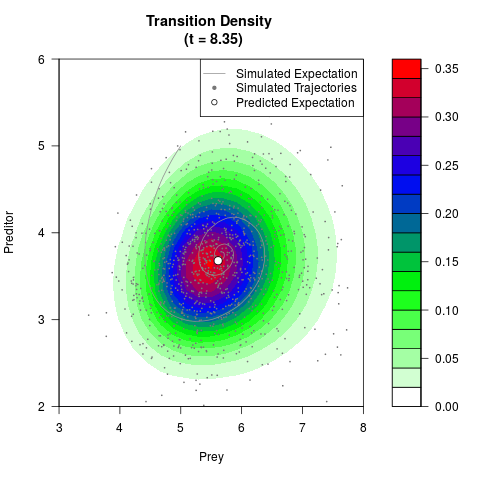
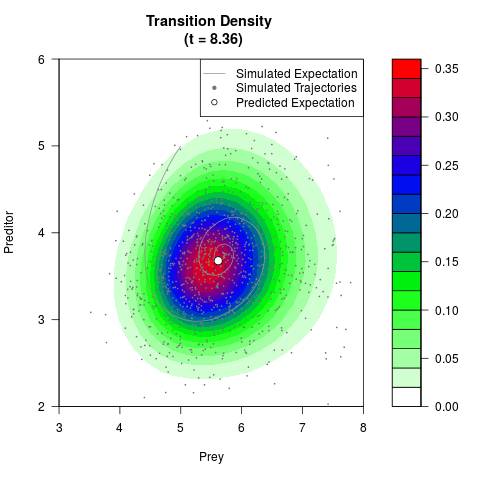
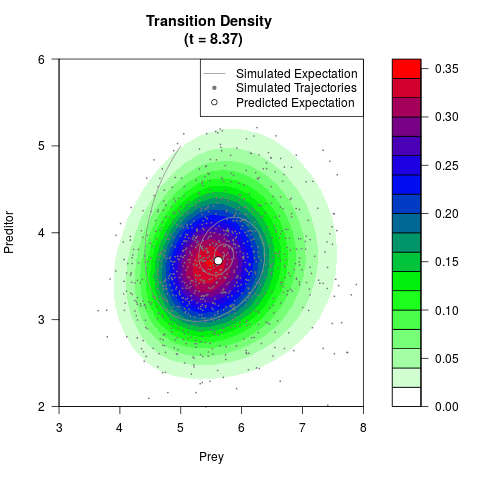
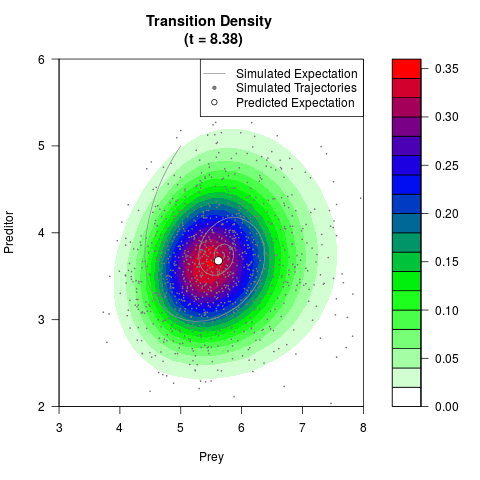
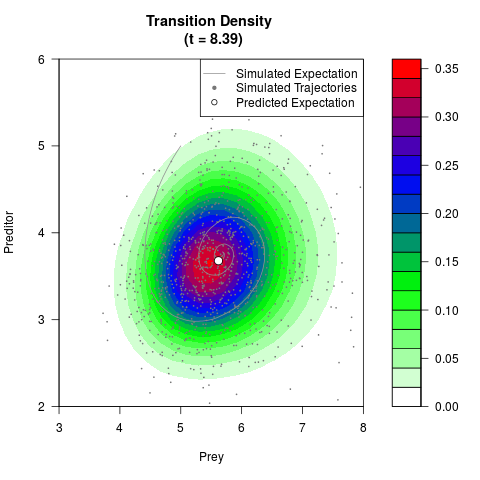
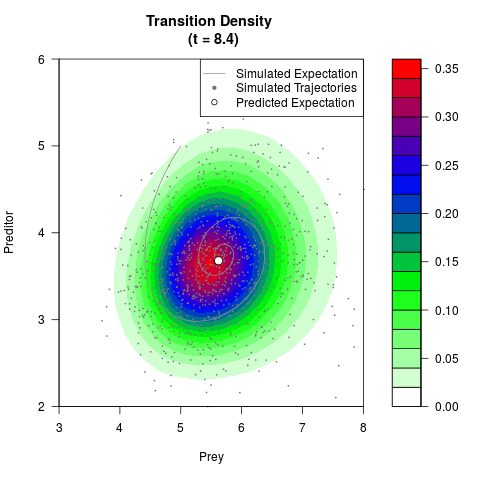
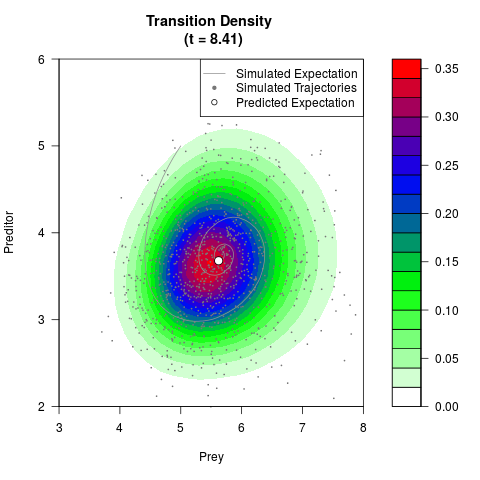
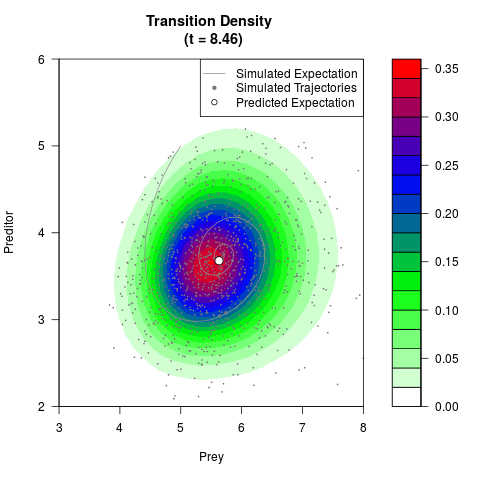
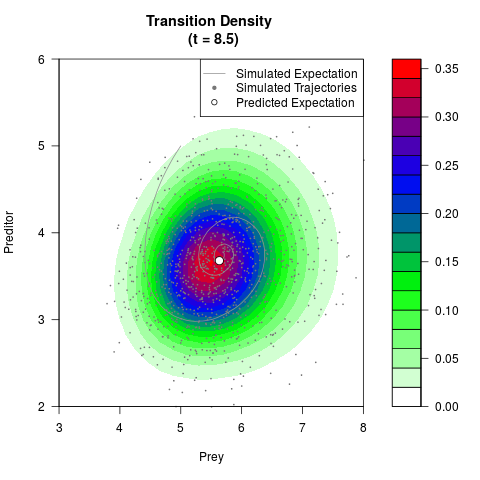
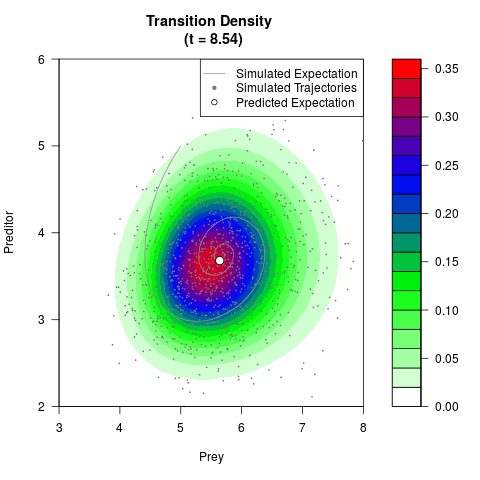
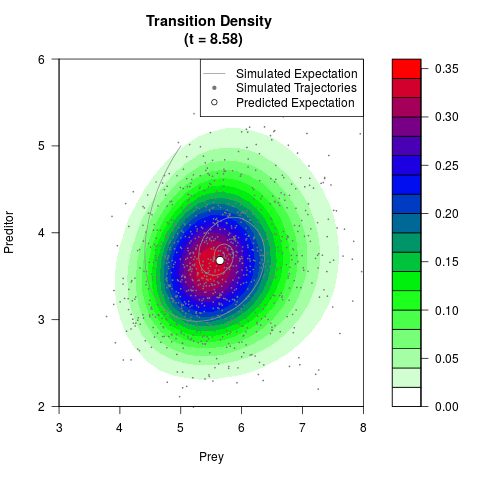
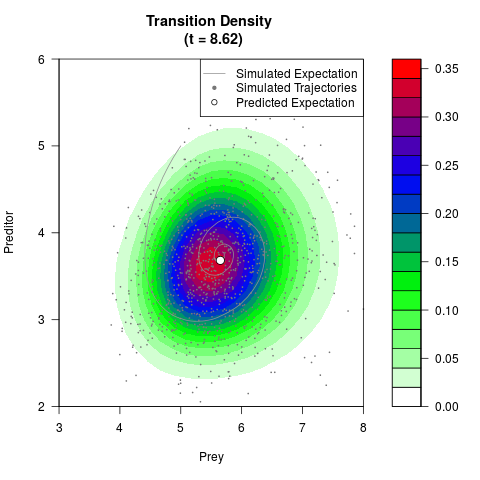
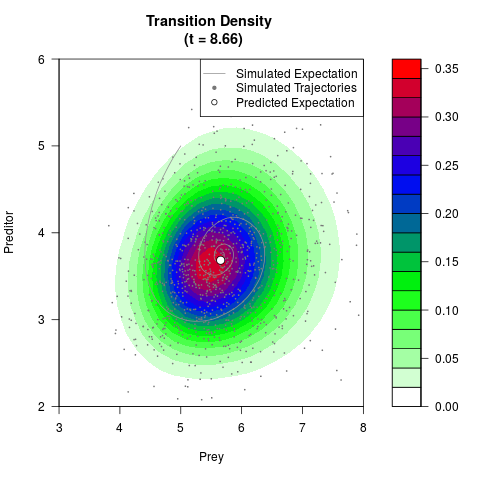
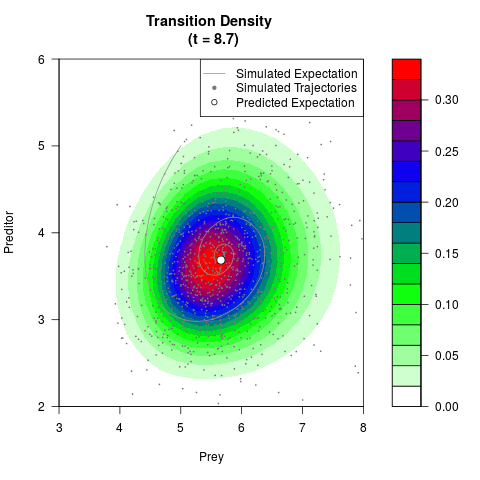
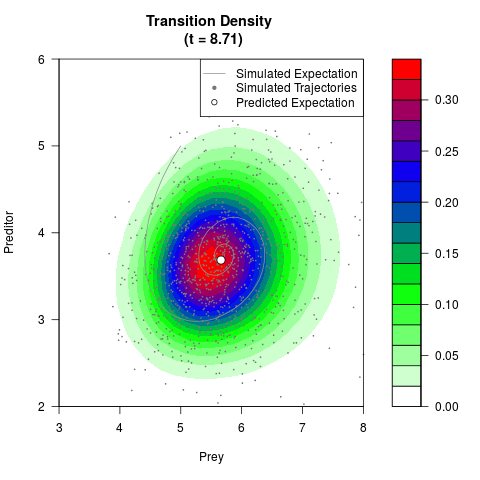
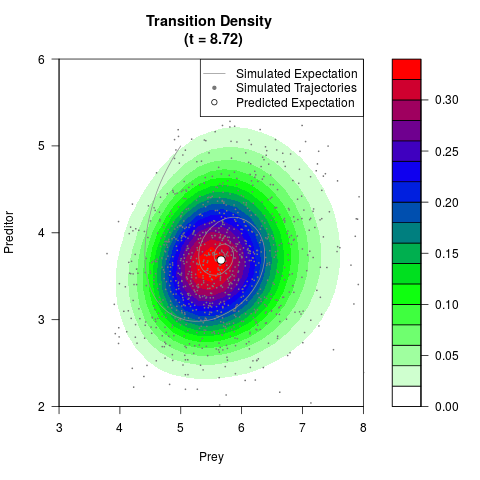
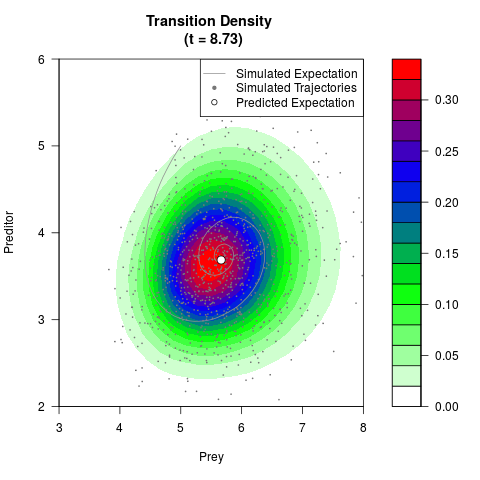
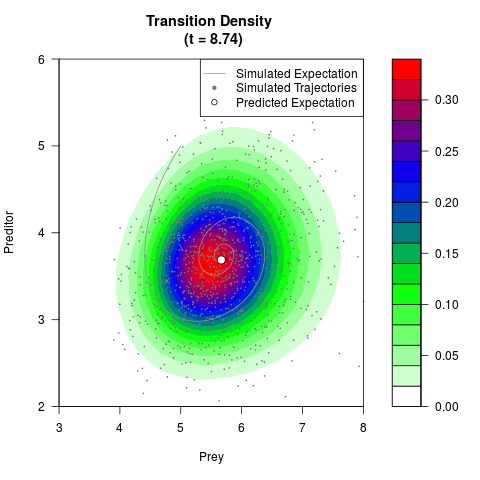
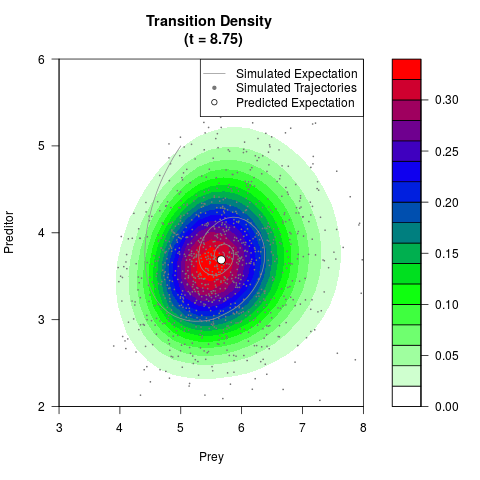
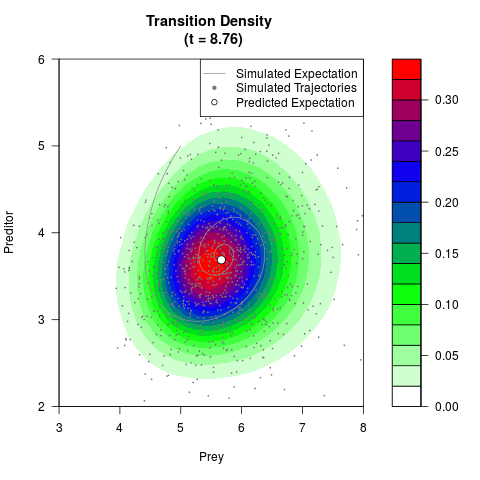
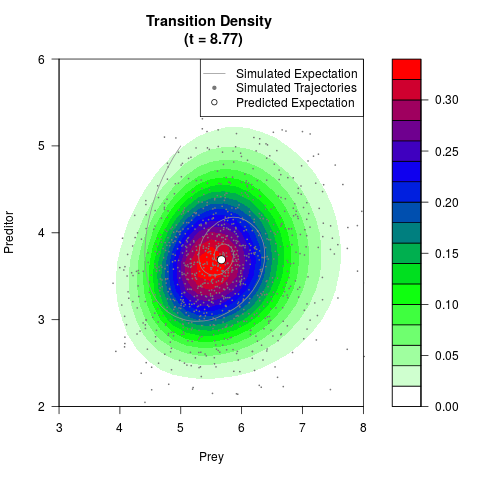
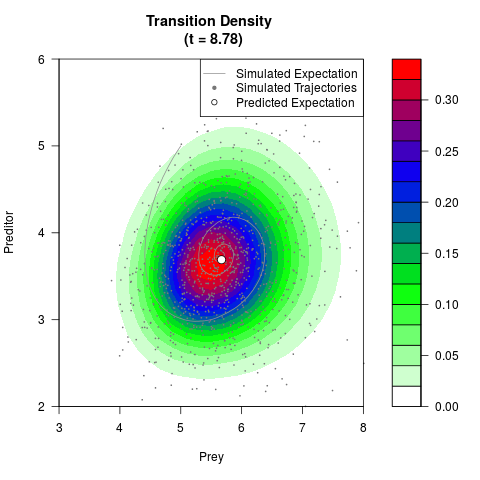
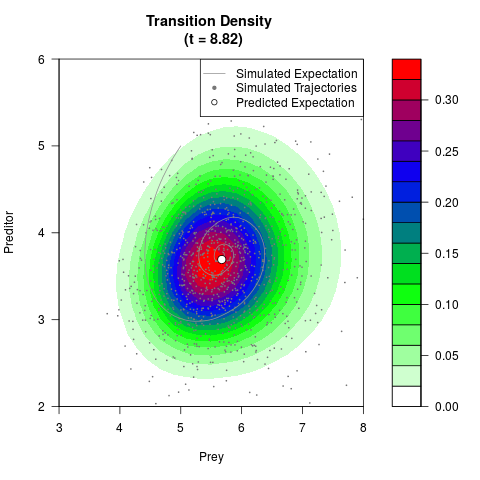
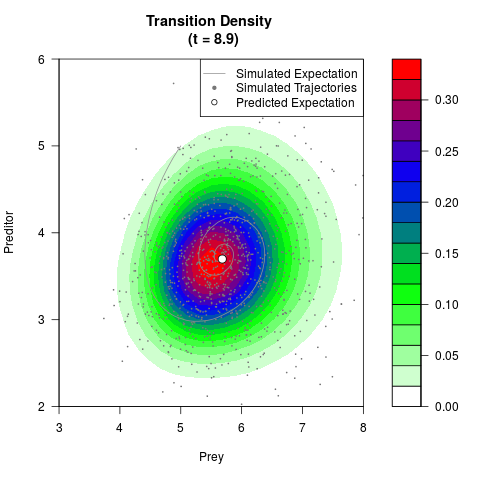
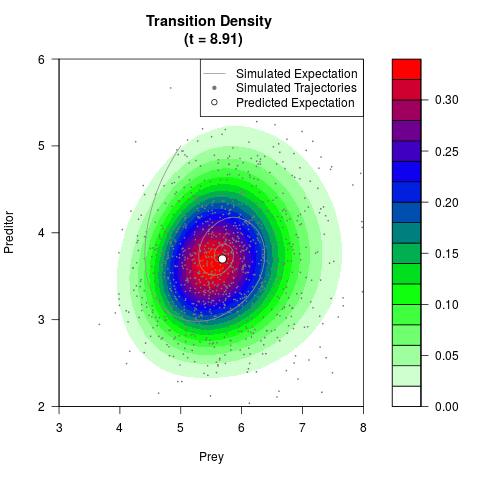
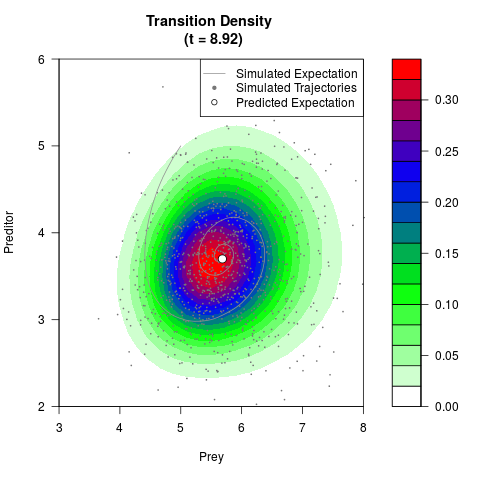
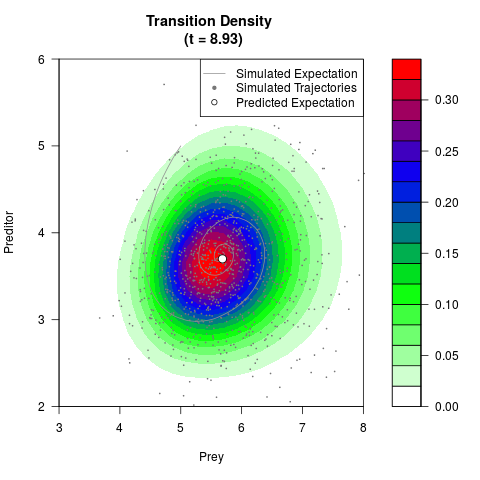
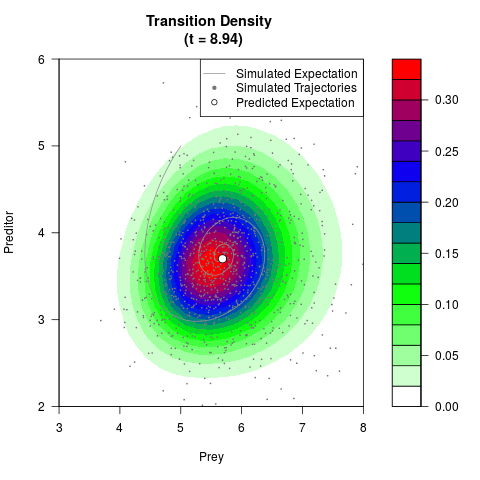
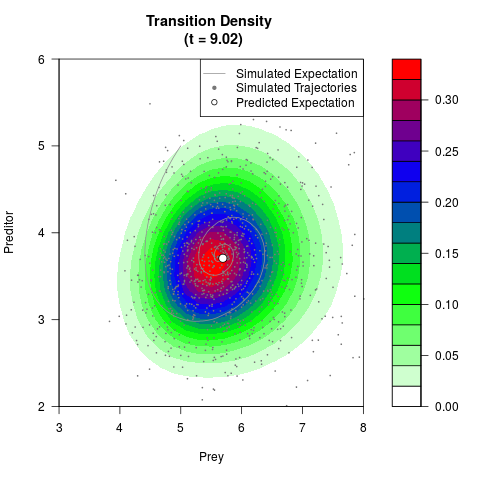
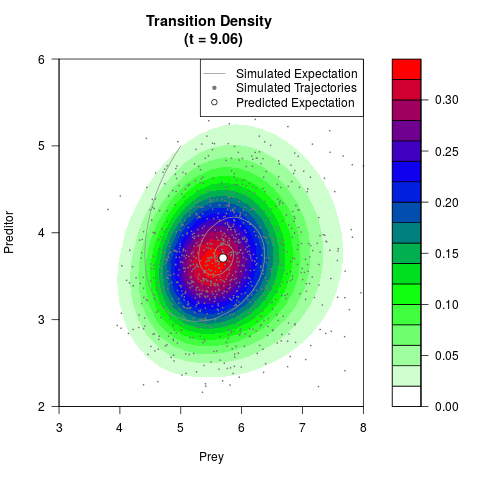
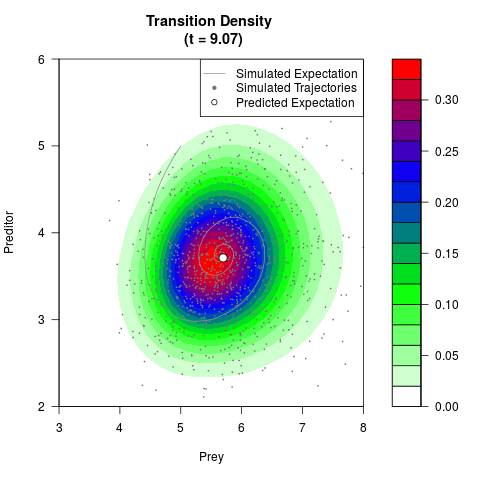
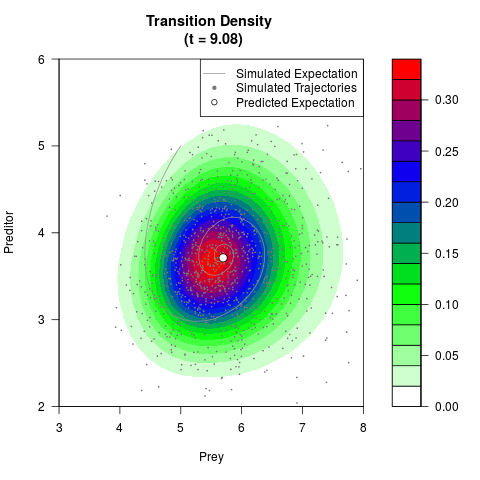
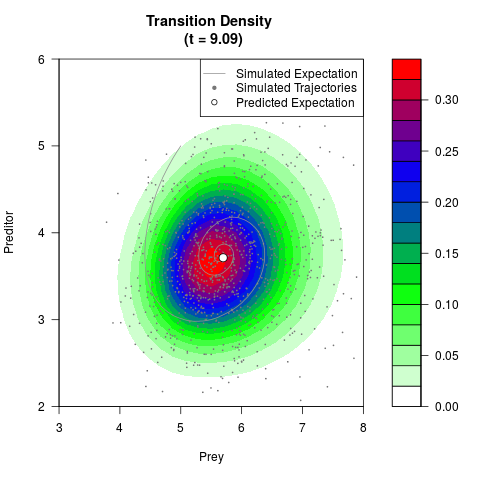
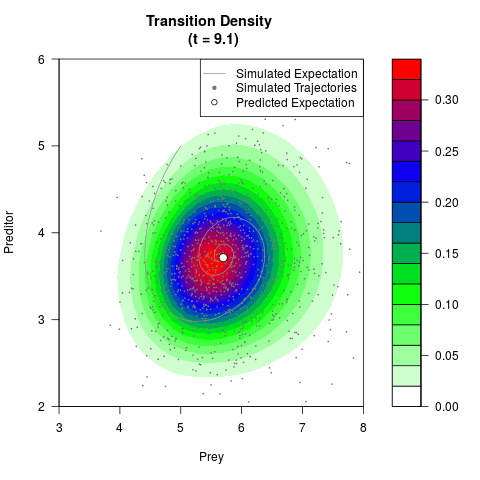
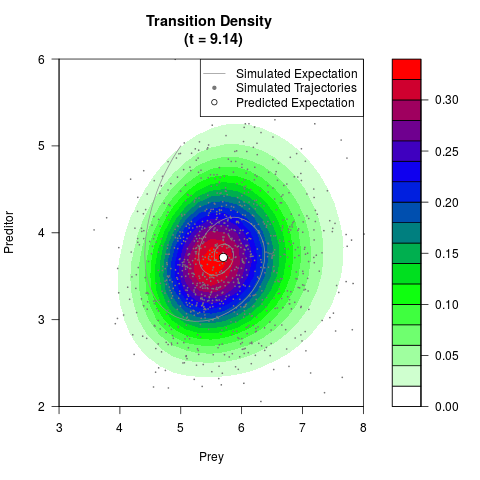
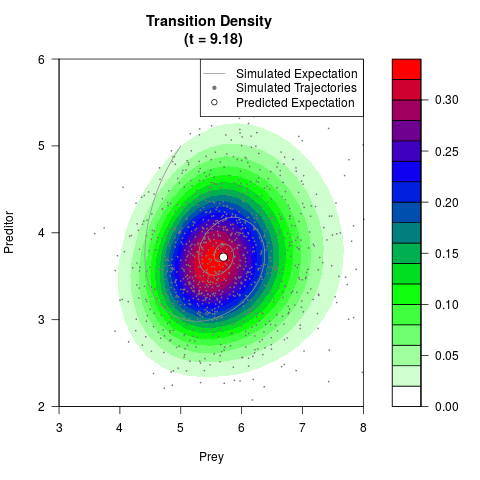
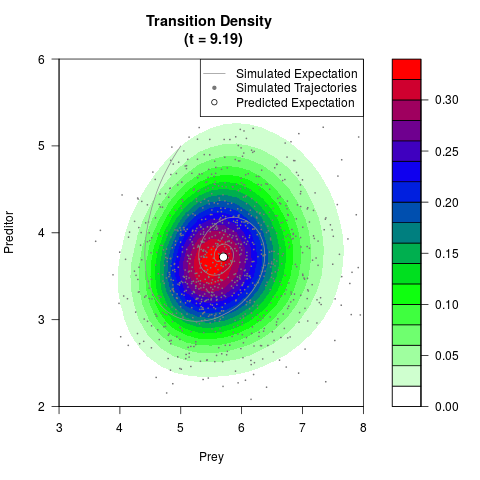
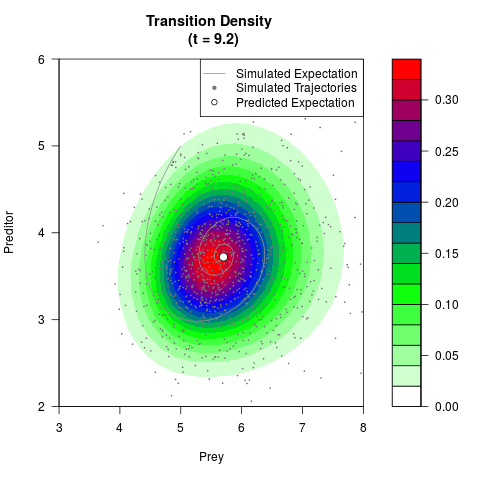
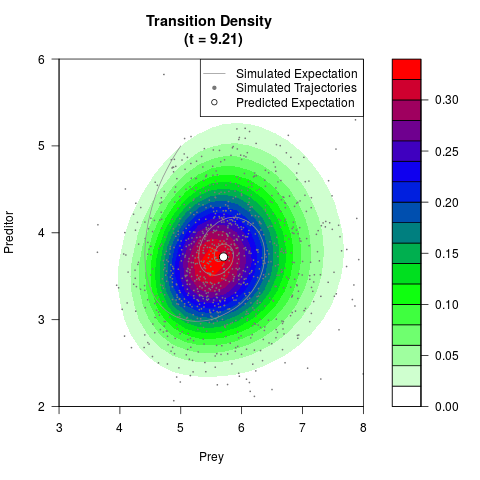
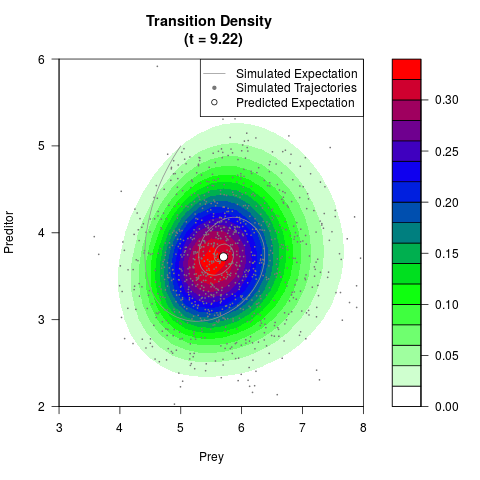
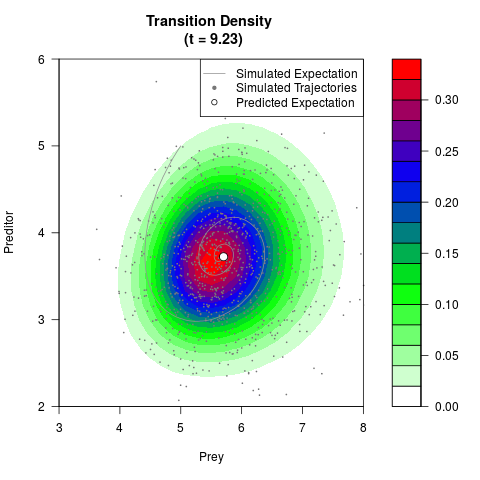
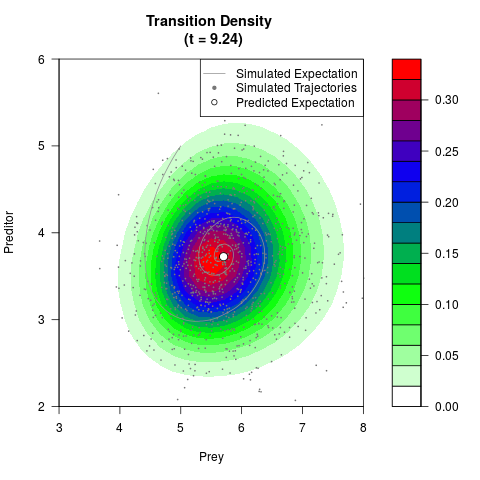
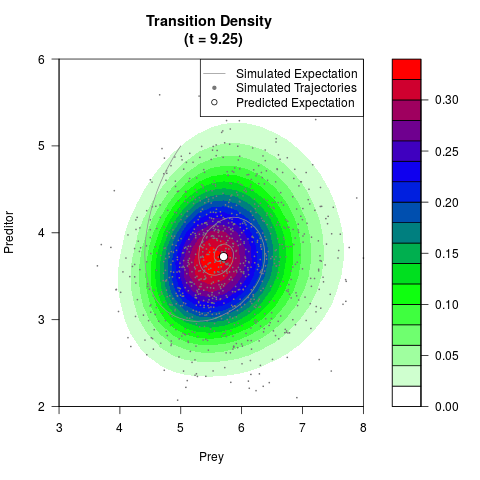
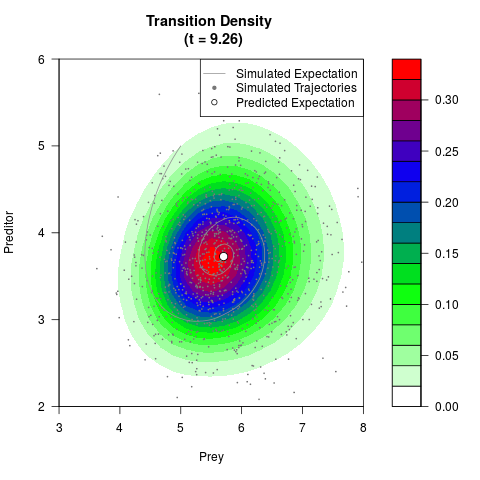
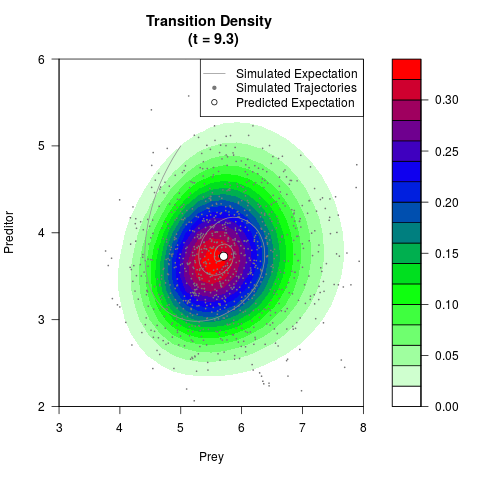
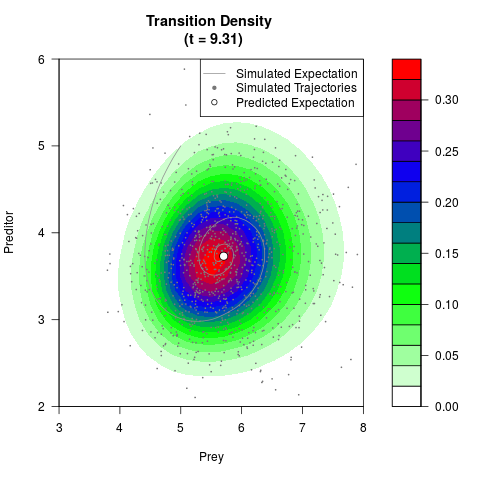
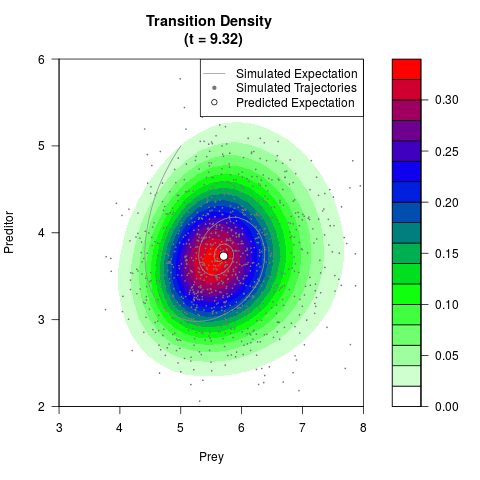
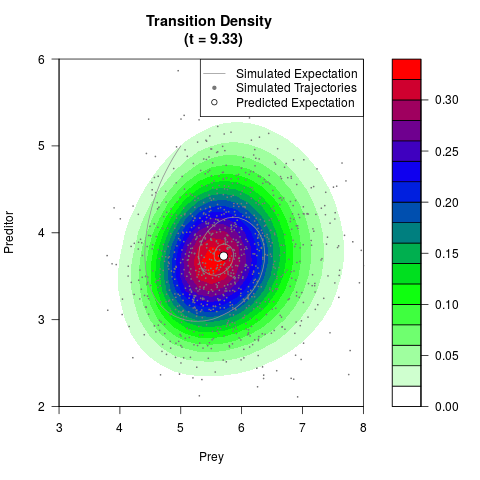
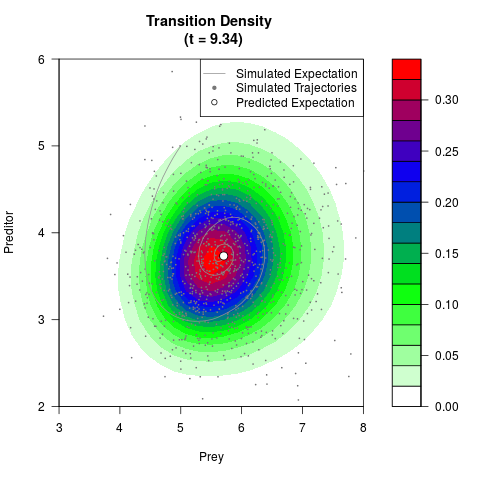
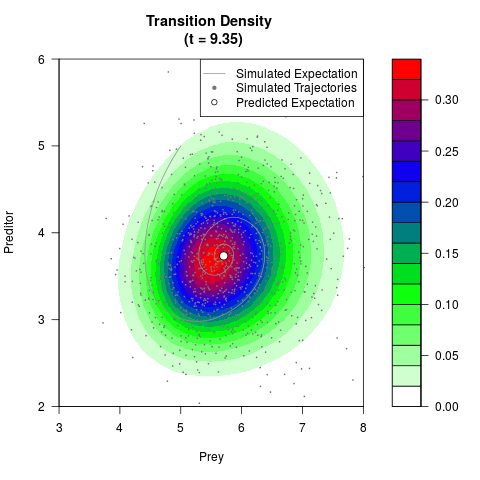
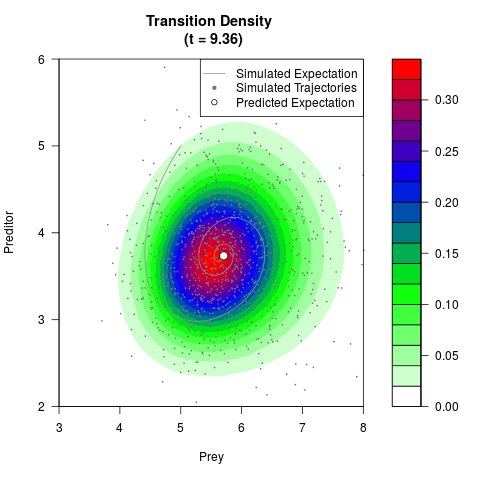
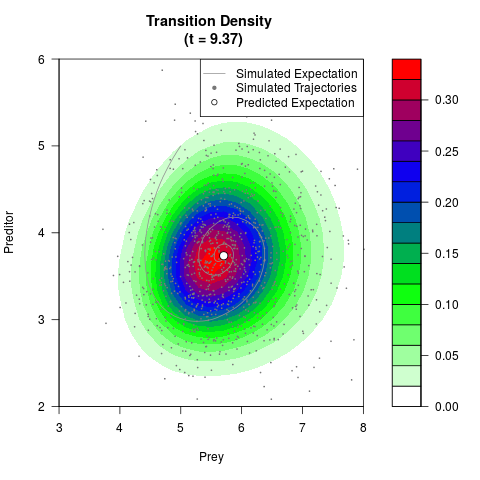
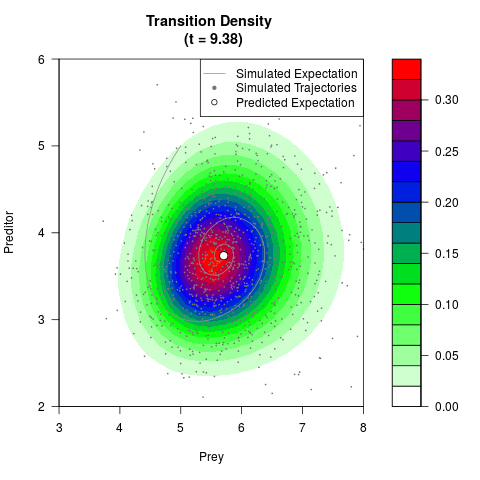
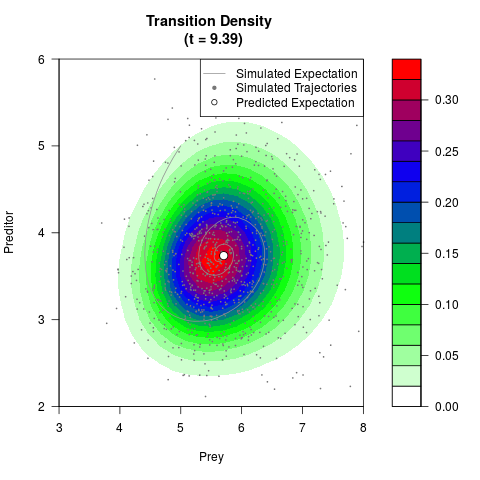
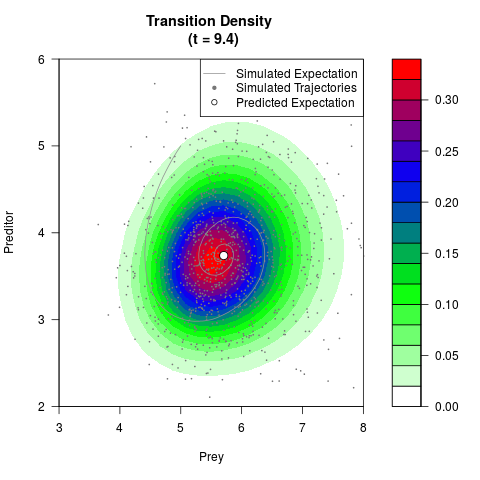
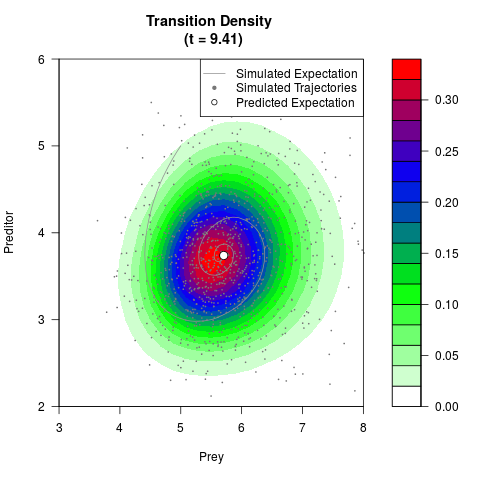
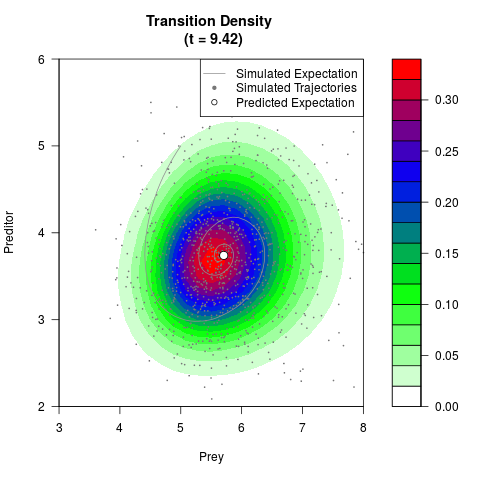
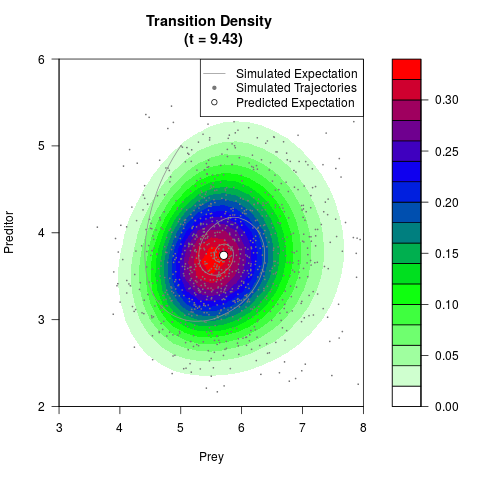
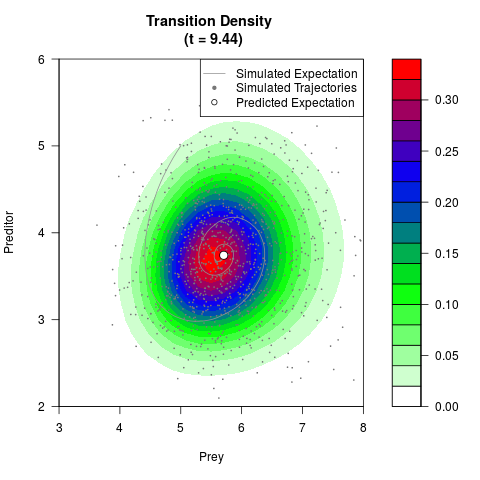
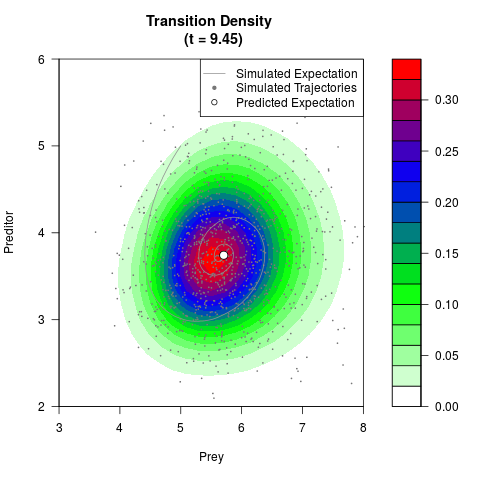
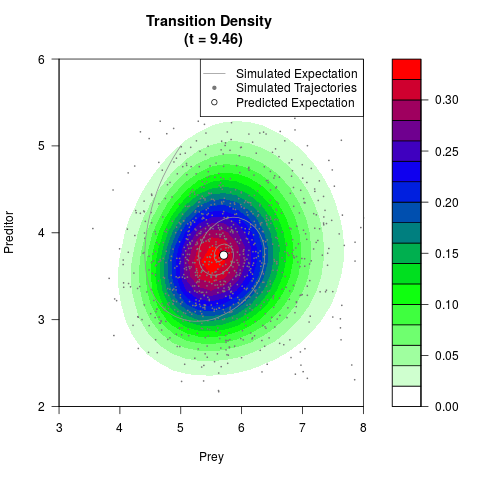
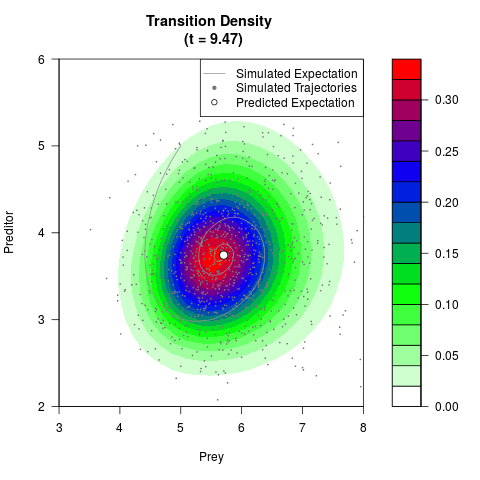
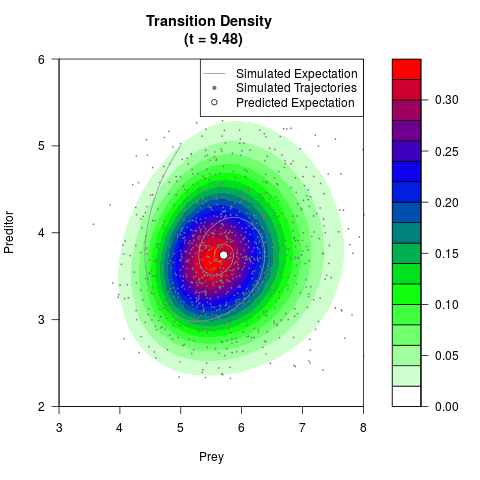
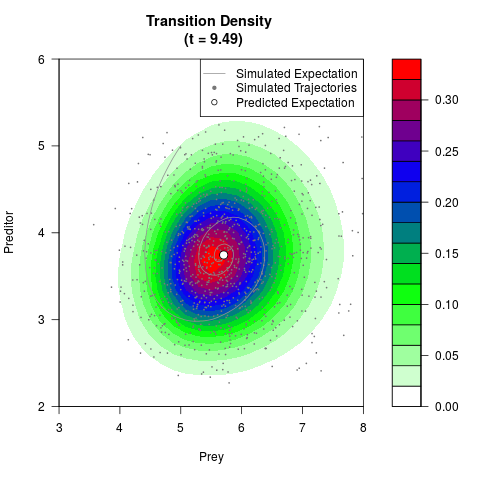
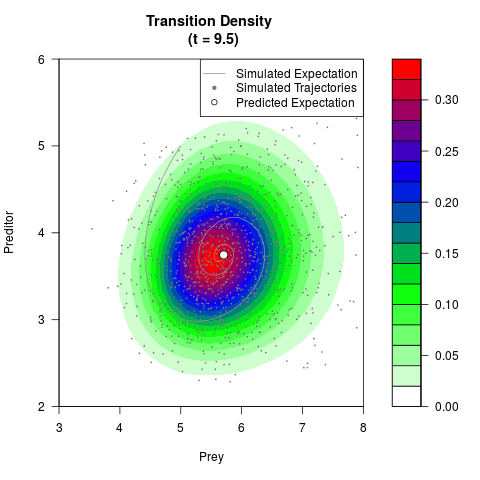
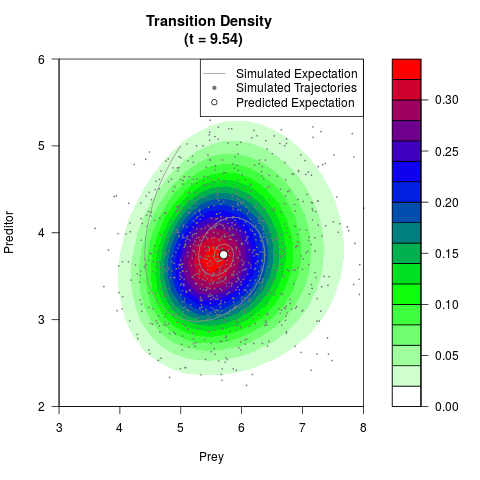
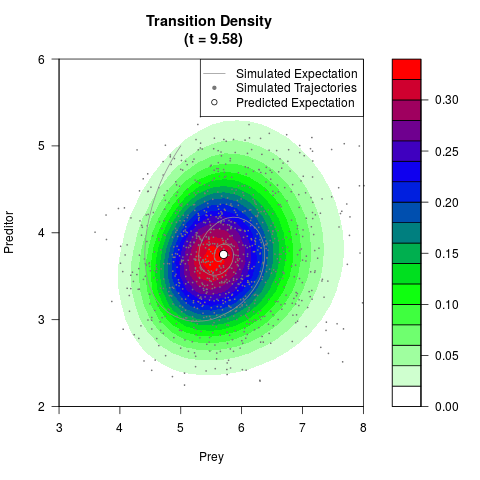
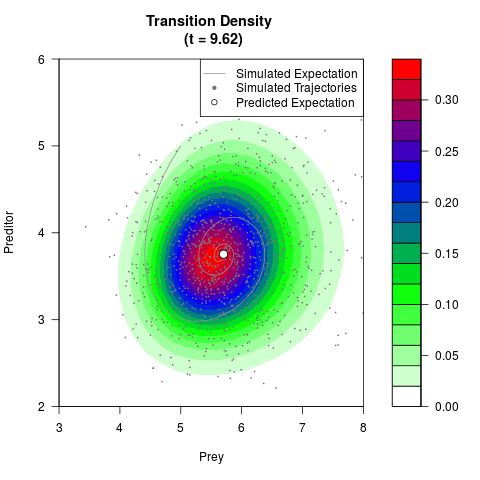
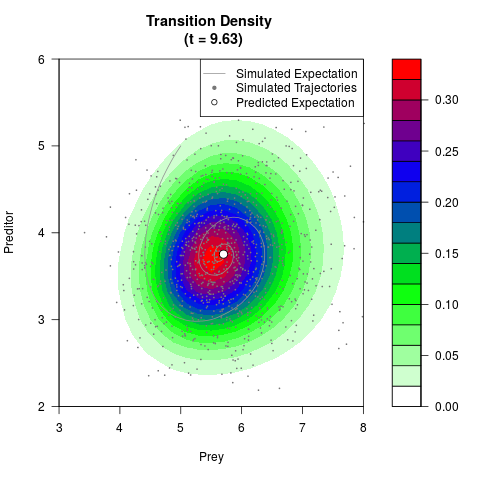
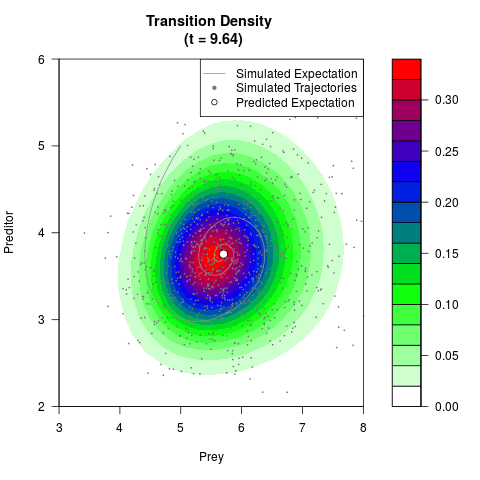
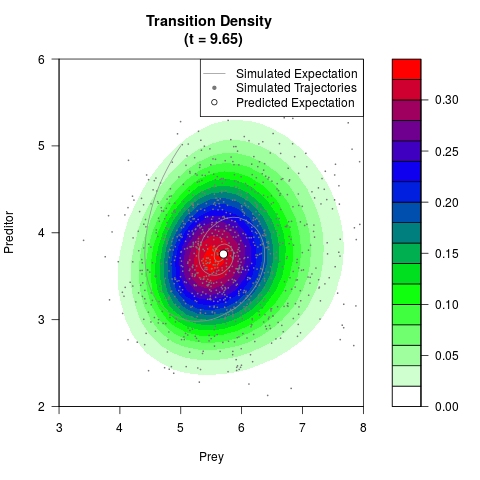
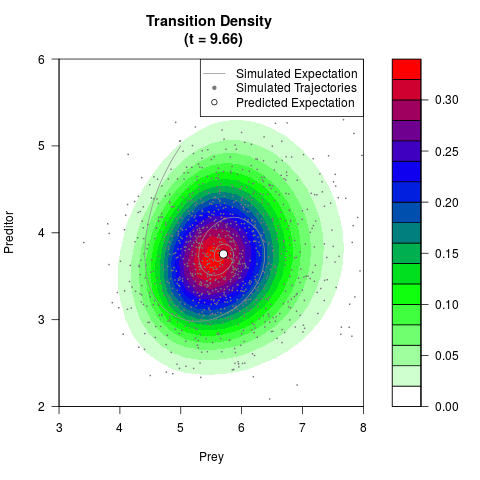
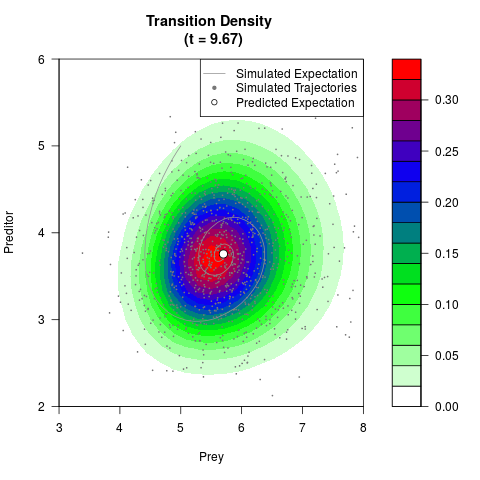
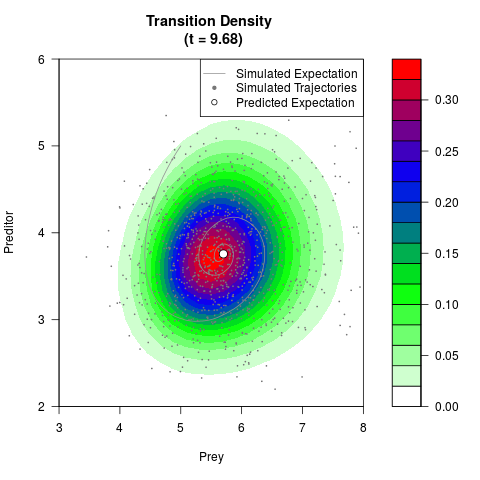
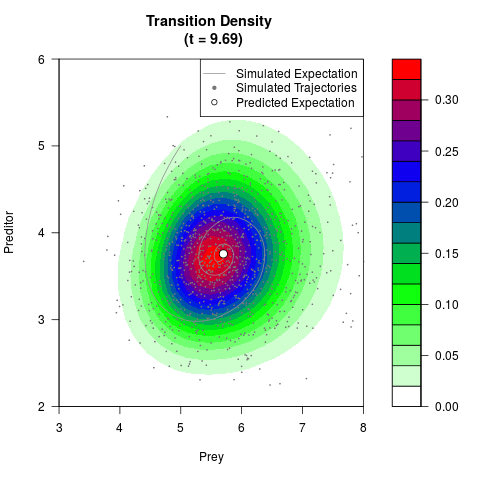
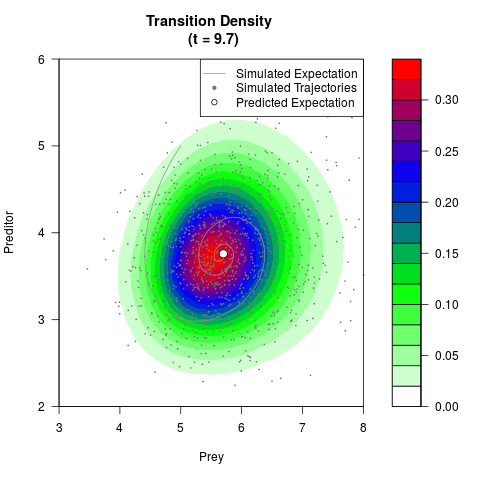
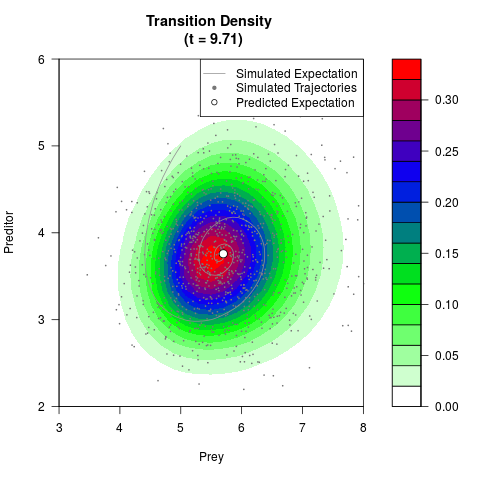
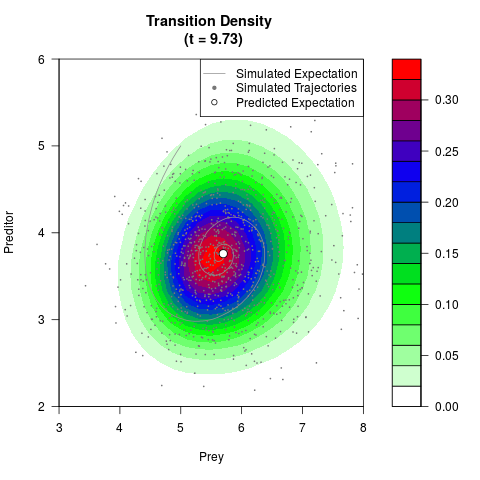
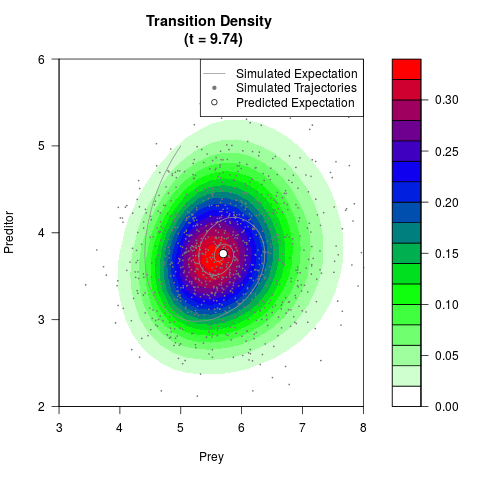
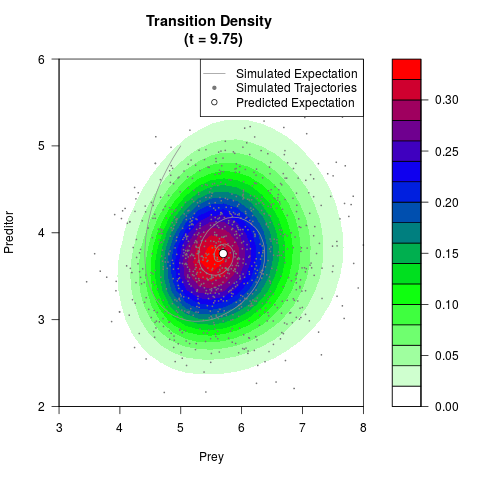
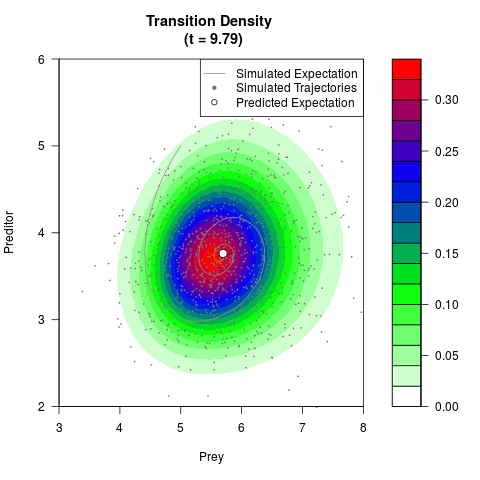
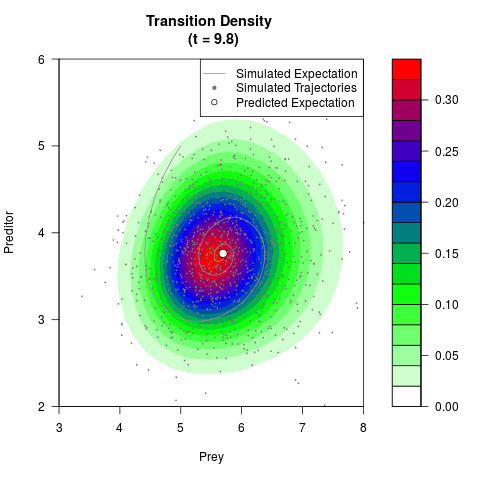
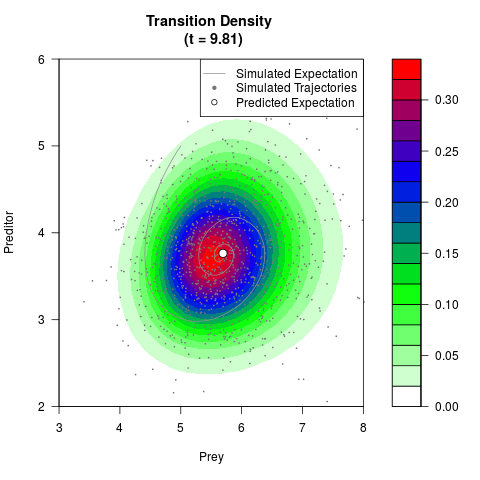
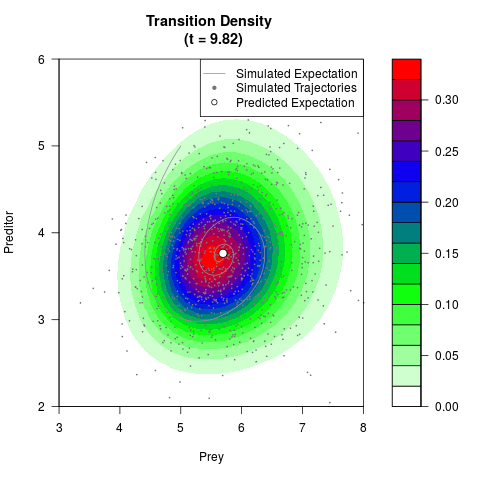
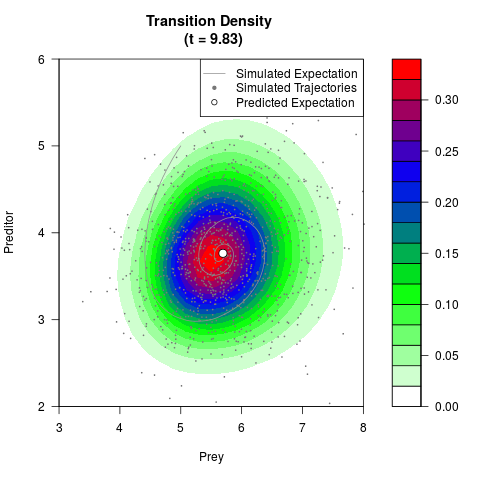
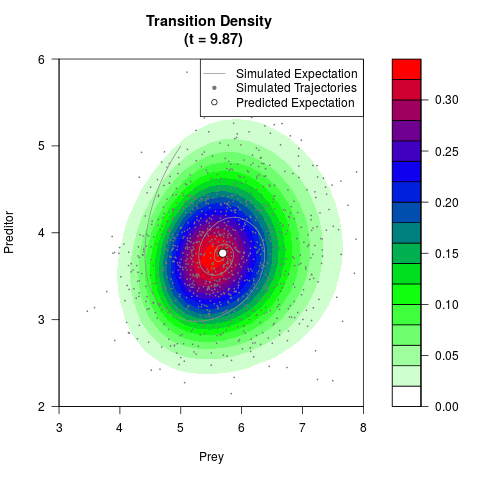
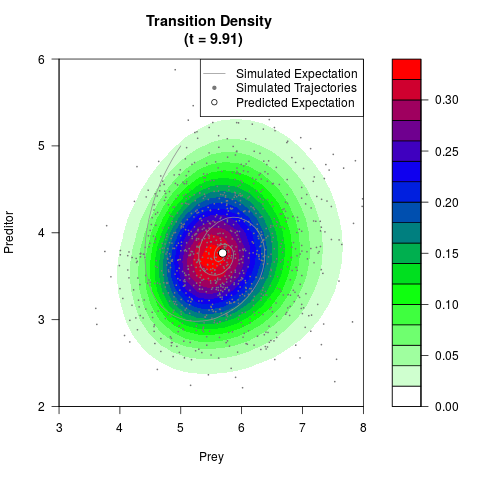
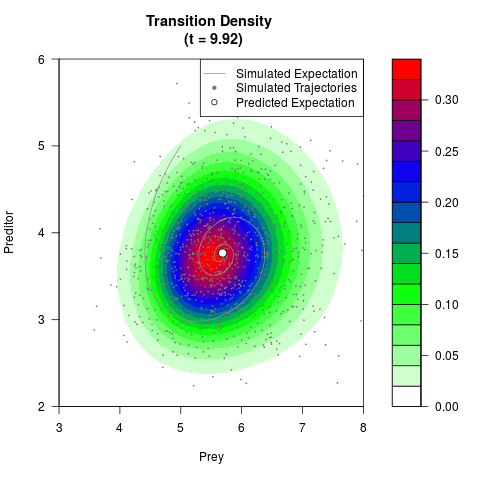
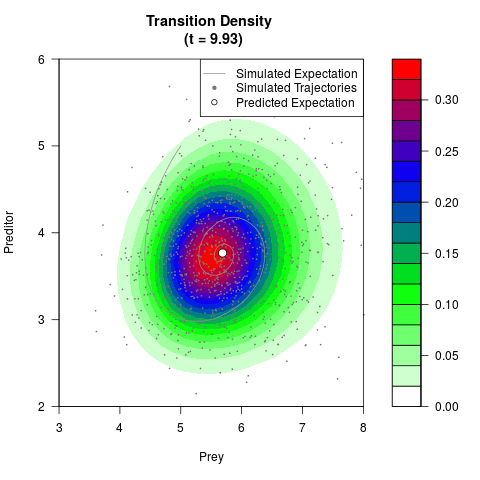
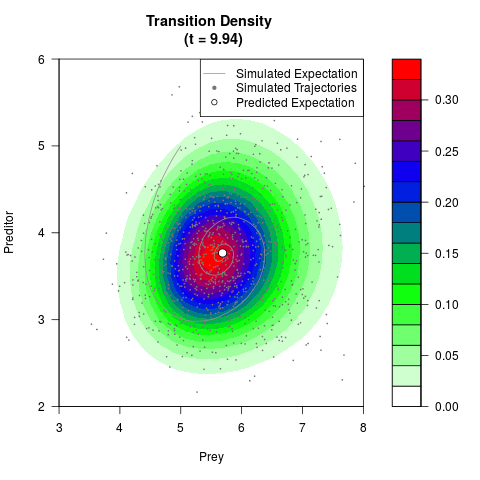
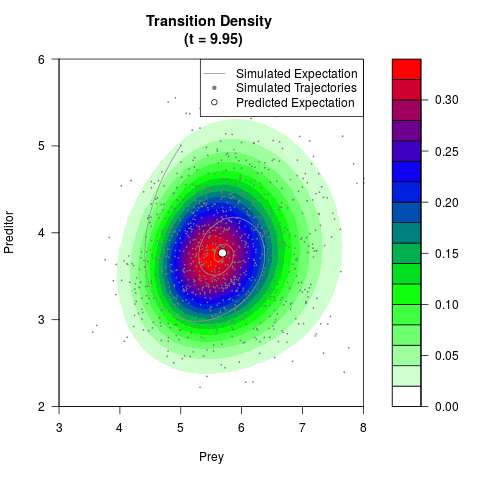
filled.contour(res$Xt,res$Yt,res$density[,,i],
main=paste0('Transition Density \n (t = ',res$time[i],')'),
color.palette=colorRampPalette(c('white','green','blue','red'))
,xlab='Prey',ylab='Preditor',plot.axes=
{
# Add simulated trajectories
points(X2~X1,pch=20,col='grey47',cex=0.01)
# Add trajectory of simulated expectation
lines(my~mx,col='grey57')
# Show the predicted expectation from BiGQD.density()
points(res$cumulants[5,i]~res$cumulants[1,i],bg='white',pch=21,cex=1.5)
axis(1);axis(2);
# Add a legend
legend('topright',lty=c('solid',NA,NA),col=c('grey57','grey47','black'),
pch=c(NA,20,21),legend=c('Simulated Expectation','Simulated Trajectories'
,'Predicted Expectation'))
})
}
#===============================================================================
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(DiffusionRgqd)
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/DiffusionRgqd/BiGQD.density.Rd_%03d_medium.png", width=480, height=480)
> ### Name: BiGQD.density
> ### Title: Generate the Transition Density of a Bivariate Generalized
> ### Quadratic Diffusion Model (2D GQD).
> ### Aliases: BiGQD.density
> ### Keywords: transition density bivariate saddlepoint bivariate Edgeworth
> ### moments cumulants
>
> ### ** Examples
>
> ## No test:
> #===============================================================================
> # Generate the transition density of a stochastic perturbed Lotka-Volterra
> # preditor-prey model, with state-dependent volatility:
> # dX = (1.5X-0.4*X*Y)dt +sqrt(0.05*X)dWt
> # dY = (-1.5Y+0.4*X*Y-0.2*Y^2)dt +sqrt(0.10*Y)dBt
> #-------------------------------------------------------------------------------
>
> # Remove any existing coefficients
> GQD.remove()
[1] "Removed : NA "
>
> # Define the X dimesnion coefficients
> a10 = function(t){1.5}
> a11 = function(t){-0.4}
> c10 = function(t){0.05}
>
> # Define the Y dimension coefficients
> b01 = function(t){-1.5}
> b11 = function(t){0.4}
> b02 = function(t){-0.2}
> f01 = function(t){0.1}
>
> # Approximate the transition density
> res = BiGQD.density(5,5,seq(3,8,length=25),seq(2,6,length=25),0,10,1/100)
================================================================
GENERALIZED QUADRATIC DIFFUSON
================================================================
_____________________ Drift Coefficients _______________________
a10 : 1.5
a11 : -0.4
... ... ... ... ... ... ... ... ... ... ...
b01 : -1.5
b02 : -0.2
b11 : 0.4
___________________ Diffusion Coefficients _____________________
c10 : 0.05
... ... ... ... ... ... ... ... ... ... ...
... ... ... ... ... ... ... ... ... ... ...
... ... ... ... ... ... ... ... ... ... ...
f01 : 0.1
=================================================================
>
>
> #------------------------------------------------------------------------------
> # Visuallize the density
> #------------------------------------------------------------------------------
>
> par(ask=FALSE)
> # Load simulated trajectory of the joint expectation:
> data(SDEsim3)
> attach(SDEsim3)
>
> # We will simulate some trajectories (crudely) as well:
> N=1000; delt= 1/100 # 1000 trajectories
> X1=rep(5,N) # Initial values for each trajectory
> X2=rep(5,N)
>
> for(i in 1:1001)
+ {
+ # Applly Euler-Murayama scheme to the LV-model
+ X1=pmax(X1+(a10(d)*X1+a11(d)*X1*X2)*delt+sqrt(c10(d)*X1)*rnorm(N,sd=sqrt(delt)),0)
+ X2=pmax(X2+(b01(d)*X2+b11(d)*X1*X2+b02(d)*X2^2)*delt+sqrt(f01(d)*X2)*rnorm(N,sd=sqrt(delt)),0)
+
+ # Now illustrate the density:
+ filled.contour(res$Xt,res$Yt,res$density[,,i],
+ main=paste0('Transition Density \n (t = ',res$time[i],')'),
+ color.palette=colorRampPalette(c('white','green','blue','red'))
+ ,xlab='Prey',ylab='Preditor',plot.axes=
+ {
+ # Add simulated trajectories
+ points(X2~X1,pch=20,col='grey47',cex=0.01)
+ # Add trajectory of simulated expectation
+ lines(my~mx,col='grey57')
+ # Show the predicted expectation from BiGQD.density()
+ points(res$cumulants[5,i]~res$cumulants[1,i],bg='white',pch=21,cex=1.5)
+ axis(1);axis(2);
+ # Add a legend
+ legend('topright',lty=c('solid',NA,NA),col=c('grey57','grey47','black'),
+ pch=c(NA,20,21),legend=c('Simulated Expectation','Simulated Trajectories'
+ ,'Predicted Expectation'))
+ })
+ }
>
> #===============================================================================
> ## End(No test)
>
>
>
>
>
>
> dev.off()
null device
1
>
|