|
Last data update: 2014.03.03
|
R: Functions for integration for Bayesian loss methodology
| tradeoff linear | R Documentation |
Functions for integration for Bayesian loss methodology
Description
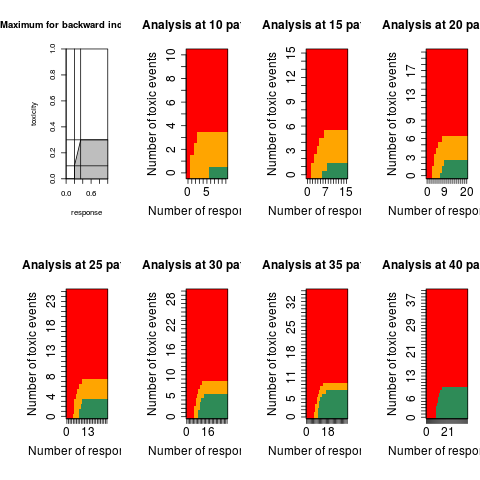
An integral and graph for an acceptable region for the bayesian loss function approach (see bayes_binom_two_loss)
Usage
tradeoff_linear_integrate(ar, br, at, bt, efficacy_region_min,
toxicity_region_max, efficacy_region_max, toxicity_region_min)
tradeoff_linear_graph(input)
Arguments
ar, br |
Parameters for the posterior distribution for response
|
at, bt |
Parameters for the posterior distribution for toxicity
|
efficacy_region_min |
Smallest acceptable efficacy
|
toxicity_region_max |
Largest acceptable toxicity
|
efficacy_region_max |
Point where no more tradeoff occurs for efficacy
|
toxicity_region_min |
Point where no more tradeoff occurs for toxicity
|
input |
A list values needed for the graph. It is expecting max.patients, efficacy_region_min, toxicity_region_max and will error without
|
Value
Returns value of the integration.
References
Chen Y, Smith BJ. Adaptive group sequential design for phase II clinical trials: a Bayesian decision theoretic approach. Stat Med 2009; 28: 3347-3362.
See Also
bayes_binom_two_loss
Integration functions and corresponding graphs:
tradeoff_square_integrate,tradeoff_ellipse_integrate,tradeoff_linear_integrate,tradeoff_ratio_integrate
Examples
# modelled toxicity probability
t=c(0.1,0.1,0.3,0.3)
# modelled response probability
r=c(0.35,0.2,0.2,0.35)
reviews=c(10,15,20,25,30,35,40)
stage_after_trial=40
# uniform prior
pra=1;prb=1;pta=1;ptb=1
efficacy_critical_value=0.2
futility_critical_value=0.35
toxicity_critical_value=0.1
no_toxicity_critical_value=0.3
# alpha/beta ratio
l_alpha_beta=3
# cost of continuing compared to cost of alpha
l_alpha_c=750
efficacy_region_min=0.2
toxicity_region_max=0.3
########################################
# linear region
efficacy_region_min=0.2
efficacy_region_max=0.35
toxicity_region_min=0.1
toxicity_region_max=0.3
s=bayes_binom_two_loss(t,r,reviews,pra,prb,pta,ptb,l_alpha_beta,
l_alpha_c,stage_after_trial,fun.integrate=tradeoff_linear_integrate,
fun.graph=tradeoff_linear_graph,efficacy_critical_value,
toxicity_critical_value,futility_critical_value,
no_toxicity_critical_value,efficacy_region_min=efficacy_region_min,
toxicity_region_max=toxicity_region_max,
efficacy_region_max=efficacy_region_max,
toxicity_region_min=toxicity_region_min)
plot(s)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(EurosarcBayes)
Loading required package: shiny
Loading required package: VGAM
Loading required package: stats4
Loading required package: splines
Loading required package: data.table
Loading required package: plyr
Loading required package: clinfun
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/EurosarcBayes/tradeoff_linear.Rd_%03d_medium.png", width=480, height=480)
> ### Name: tradeoff linear
> ### Title: Functions for integration for Bayesian loss methodology
> ### Aliases: tradeoff_linear_integrate tradeoff_linear_graph
>
> ### ** Examples
>
> # modelled toxicity probability
> t=c(0.1,0.1,0.3,0.3)
> # modelled response probability
> r=c(0.35,0.2,0.2,0.35)
>
> reviews=c(10,15,20,25,30,35,40)
> stage_after_trial=40
>
> # uniform prior
> pra=1;prb=1;pta=1;ptb=1
>
> efficacy_critical_value=0.2
> futility_critical_value=0.35
> toxicity_critical_value=0.1
> no_toxicity_critical_value=0.3
>
> # alpha/beta ratio
> l_alpha_beta=3
> # cost of continuing compared to cost of alpha
> l_alpha_c=750
>
> efficacy_region_min=0.2
> toxicity_region_max=0.3
>
> ########################################
> # linear region
> efficacy_region_min=0.2
> efficacy_region_max=0.35
> toxicity_region_min=0.1
> toxicity_region_max=0.3
>
> s=bayes_binom_two_loss(t,r,reviews,pra,prb,pta,ptb,l_alpha_beta,
+ l_alpha_c,stage_after_trial,fun.integrate=tradeoff_linear_integrate,
+ fun.graph=tradeoff_linear_graph,efficacy_critical_value,
+ toxicity_critical_value,futility_critical_value,
+ no_toxicity_critical_value,efficacy_region_min=efficacy_region_min,
+ toxicity_region_max=toxicity_region_max,
+ efficacy_region_max=efficacy_region_max,
+ toxicity_region_min=toxicity_region_min)
[1] "The cost function is constant for all patients"
cut-points at each analysis
patient review low toxicity high toxicity poor outcome good outcome
1 10 0 4 0 6
2 15 1 6 1 6
3 20 2 7 2 7
4 25 3 8 3 8
5 30 5 9 5 9
6 35 7 10 6 10
7 40 9 10 9 10
Frequentist properties of design
Stopping rules T=0.1, R=0.35 T=0.1, R=0.2
1 Stop early - Futility/Toxicity 20.40 73.02
4 Continue to final analysis - Futility/Toxicity 4.47 8.53
2 Stop early - Efficacy 67.90 12.71
3 Continue to final analysis - Efficacy 7.22 5.74
6 Expected number of patients recruited 24.91 23.31
T=0.3, R=0.2 T=0.3, R=0.35
1 97.55 85.59
4 1.03 4.18
2 1.00 6.75
3 0.42 3.48
6 13.80 18.33
Bayesian properties of trial design
n T>0.3 T>0.1 T>0.3 T>0.1 R>0.2 R>0.35 R>0.2 R>0.35
10 0.020 0.314 0.790 0.997 0.086 0.009 0.998 0.950
15 0.026 0.515 0.825 0.999 0.141 0.010 0.973 0.688
20 0.027 0.648 0.723 0.999 0.179 0.009 0.957 0.536
25 0.026 0.741 0.627 0.999 0.207 0.007 0.941 0.411
30 0.063 0.917 0.542 0.999 0.393 0.018 0.925 0.311
35 0.112 0.976 0.466 0.999 0.401 0.013 0.911 0.234
40 0.170 0.994 0.275 0.998 0.704 0.052 0.818 0.102
Futility P(R<0.35)=0.948
Efficacy P(R>0.2)=0.818
Toxicity ok P(T<0.3)=0.83
Toxicity P(T>0.1)=0.997>
> plot(s)
>
>
>
>
>
>
>
> dev.off()
null device
1
>
|
|
 providing
providing  .
.