Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Computes the Leslie or Delury population estimate from catch and effort data.DescriptionComputes the Leslie or Delury estimates of population size and catchability coefficient from paired catch and effort data. The Ricker modification may also be used. Usage
depletion(catch, effort, method = c("Leslie", "DeLury", "Delury"),
Ricker.mod = FALSE)
## S3 method for class 'depletion'
summary(object, type = c("params", "lm"),
verbose = FALSE, digits = getOption("digits"), ...)
## S3 method for class 'depletion'
coef(object, type = c("params", "lm"),
digits = getOption("digits"), ...)
## S3 method for class 'depletion'
confint(object, parm = c("No", "q", "lm"),
level = conf.level, conf.level = 0.95, digits = getOption("digits"),
...)
## S3 method for class 'depletion'
anova(object, ...)
## S3 method for class 'depletion'
plot(x, xlab = NULL, ylab = NULL, pch = 19,
col.pt = "black", col.mdl = "gray70", lwd = 1, lty = 1,
pos.est = "topright", cex.est = 0.95, ...)
Arguments
DetailsFor the Leslie method, a linear regression model of catch-per-unit-effort on cumulative catch prior to the sample is fit. The catchability coefficient (q) is estimated from the negative of the slope and the initial population size (No) is estimated by dividing the intercept by the catchability coefficient. If For the DeLury method, a linear regression model of log (catch-per-unit-effort) on cumulative effort is fit. The catchability coefficient (q) is estimated from the negative of the slope and the initial population size (No) is estimated by dividing the intercept as an exponent of e by the catchability coefficient. If Standard errors for the catchability and population size estimates are computed fronm formulas on page 298 (for Leslie) and 303 (for DeLury) from Seber (2002). Confidence intervals are computed using standard large-sample normal distribution theory with the regression error df. ValueA list with the following items:
testingThe Leslie method without the Ricker modification and the DeLury method with the Ricker modification matches the results from The Leslie method matches the results of Seber (2002) for N0, q, and the CI for Q but not the CI for N (which was so far off that it might be that Seber's result is incorrect) for the lobster data and the q and CI for q but the NO or its CI (likely due to lots of rounding in Seber 2002) for the Blue Crab data. The Leslie and DeLury methods match the results of Ricker (1975) for No and Q but not for the CI of No (Ricker used a very different method to compute CIs). IFAR Chapter10-Abundance from Depletion Data. Author(s)Derek H. Ogle, derek@derekogle.com ReferencesOgle, D.H. 2016. Introductory Fisheries Analyses with R. Chapman & Hall/CRC, Boca Raton, FL. Ricker, W.E. 1975. Computation and interpretation of biological statistics of fish populations. Technical Report Bulletin 191, Bulletin of the Fisheries Research Board of Canada. [Was (is?) from http://www.dfo-mpo.gc.ca/Library/1485.pdf.] Seber, G.A.F. 2002. The Estimation of Animal Abundance. Edward Arnold, Second edition (reprinted). See AlsoSee Examplesdata(SMBassLS) ## Leslie model examples # no Ricker modification l1 <- depletion(SMBassLS$catch,SMBassLS$effort,method="Leslie") coef(l1) summary(l1) summary(l1,verbose=TRUE) confint(l1) summary(l1,type="lm") plot(l1) # with Ricker modification l2 <- depletion(SMBassLS$catch,SMBassLS$effort,method="Leslie",Ricker.mod=TRUE) summary(l2) confint(l2) plot(l2) ## Delury model examples # no Ricker modification d1 <- depletion(SMBassLS$catch,SMBassLS$effort,method="Delury") coef(d1) summary(d1) summary(d1,verbose=TRUE) confint(d1) summary(d1,type="lm") plot(d1) # with Ricker modification d2 <- depletion(SMBassLS$catch,SMBassLS$effort,method="Delury",Ricker.mod=TRUE) summary(d2) confint(d2) plot(d2) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(FSA)
############################################
## FSA package, version 0.8.7 ##
## Derek H. Ogle, Northland College ##
## ##
## Run ?FSA for documentation. ##
## Run citation('FSA') for citation ... ##
## please cite if used in publication. ##
## ##
## See derekogle.com/fishR/ for more ##
## thorough analytical vignettes. ##
############################################
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/FSA/depletion.Rd_%03d_medium.png", width=480, height=480)
> ### Name: depletion
> ### Title: Computes the Leslie or Delury population estimate from catch and
> ### effort data.
> ### Aliases: anova.depletion coef.depletion confint.depletion depletion
> ### plot.depletion summary.depletion
> ### Keywords: hplot manip
>
> ### ** Examples
>
> data(SMBassLS)
>
> ## Leslie model examples
> # no Ricker modification
> l1 <- depletion(SMBassLS$catch,SMBassLS$effort,method="Leslie")
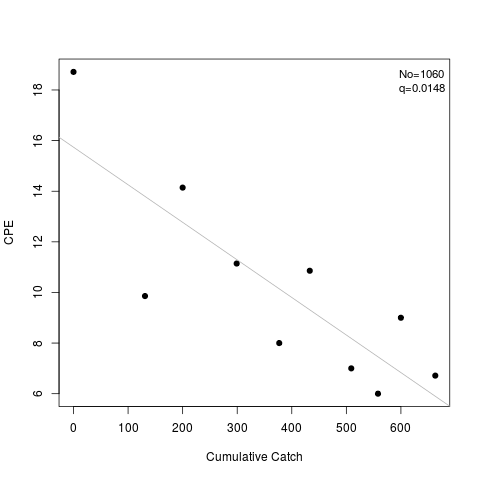
> coef(l1)
No q
[1,] 1060.296 0.014844
> summary(l1)
Estimate Std. Err.
No 1060.295599 169.2676197
q 0.014844 0.0035205
> summary(l1,verbose=TRUE)
The Leslie method was used.
Estimate Std. Err.
No 1060.295599 169.2676197
q 0.014844 0.0035205
> confint(l1)
95% LCI 95% UCI
No 669.9637682 1450.6274302
q 0.0067257 0.0229623
> summary(l1,type="lm")
Call:
stats::lm(formula = cpe ~ K)
Residuals:
Min 1Q Median 3Q Max
-3.9373 -1.3879 0.3295 1.5023 2.9752
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 15.73906 1.51266 10.405 6.31e-06 ***
K -0.01484 0.00352 -4.216 0.00293 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.295 on 8 degrees of freedom
Multiple R-squared: 0.6897, Adjusted R-squared: 0.6509
F-statistic: 17.78 on 1 and 8 DF, p-value: 0.00293
> plot(l1)
>
> # with Ricker modification
> l2 <- depletion(SMBassLS$catch,SMBassLS$effort,method="Leslie",Ricker.mod=TRUE)
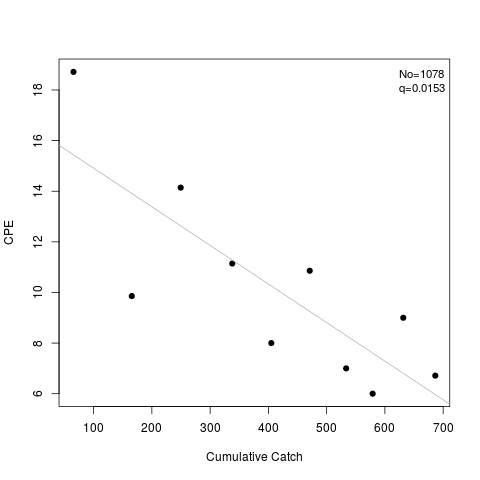
> summary(l2)
Estimate Std. Err.
No 1077.5713043 177.8035211
q 0.0152508 0.0039116
> confint(l2)
95% LCI 95% UCI
No 667.5556494 1487.586959
q 0.0062306 0.024271
> plot(l2)
>
> ## Delury model examples
> # no Ricker modification
> d1 <- depletion(SMBassLS$catch,SMBassLS$effort,method="Delury")
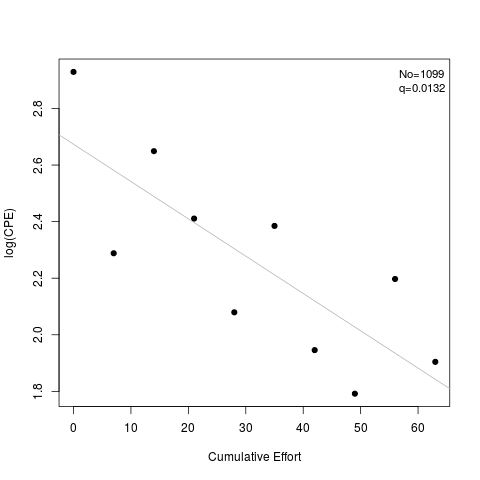
> coef(d1)
No q
[1,] 1098.503 0.0131938
> summary(d1)
Estimate Std. Err.
No 1098.5032552 191.6048926
q 0.0131938 0.0035858
> summary(d1,verbose=TRUE)
The DeLury method was used.
Estimate Std. Err.
No 1098.5032552 191.6048926
q 0.0131938 0.0035858
> confint(d1)
95% LCI 95% UCI
No 656.6615805 1540.3449299
q 0.0049249 0.0214627
> summary(d1,type="lm")
Call:
stats::lm(formula = log(cpe) ~ E)
Residuals:
Min 1Q Median 3Q Max
-0.29314 -0.21203 0.03796 0.16974 0.26238
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.673692 0.134000 19.953 4.15e-08 ***
E -0.013194 0.003586 -3.679 0.00622 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.228 on 8 degrees of freedom
Multiple R-squared: 0.6286, Adjusted R-squared: 0.5821
F-statistic: 13.54 on 1 and 8 DF, p-value: 0.006224
> plot(d1)
>
> # with Ricker modification
> d2 <- depletion(SMBassLS$catch,SMBassLS$effort,method="Delury",Ricker.mod=TRUE)
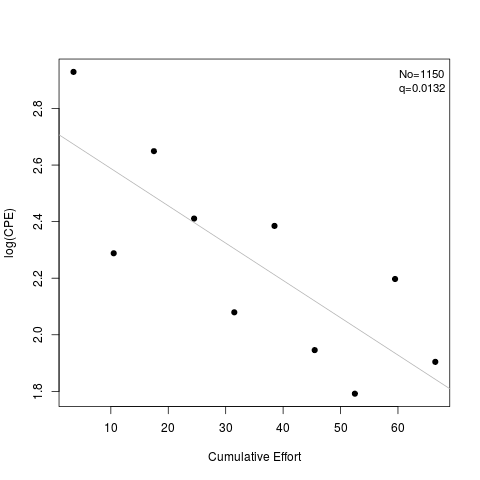
> summary(d2)
Estimate Std. Err.
No 1.15042e+03 187.6083109
q 1.31938e-02 0.0035858
> confint(d2)
95% LCI 95% UCI
No 717.7940216 1583.0451030
q 0.0049249 0.0214627
> plot(d2)
>
>
>
>
>
>
> dev.off()
null device
1
>
|