Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Abbreviating items (from questionnaire or other) measures using Genetic Algorithms (GAs)DescriptionThe GAabbreviate uses Genetic Algorithms as an optimization tool for scale abbreviation or subset selection that maximally captures the variance in the original data. Usage
GAabbreviate(items = NULL,
scales = NULL,
itemCost = 0.05,
maxItems = 5,
maxiter = 100,
popSize = 50,
...,
plot = FALSE,
verbose = interactive(),
crossVal = TRUE,
impute = FALSE,
pairwise = FALSE,
minR = 0,
sWeights = NULL,
nSample = NULL,
seed = NULL)
Arguments
DetailsThe GAabbreviate uses Genetic Algorithms (GAs) as an optimization tool for shortening a large set of variables (e.g., in a lengthy battery of questionnaires) into a shorter subset that maximally captures the variance in the original data. An exhaustive search of all possible shorter forms of the original measure would be time consuming, especially for a measure with a large number of items. For a long form of length L (e.g., 100 items of a self-report scale), the size of the search space is 2^L (1.26e+30) and forms a hypercube of L dimensions. The GA uses hypercube sampling by sampling the corners of the L-dimensional hypercube. It optimizes the search by mimicking Darwinian evolution mechanisms (of selection, crossover, and mutation) while searching through a "landscape" of the collection of all possible fitness values to find an optimal value. This does not imply that the GA finds the "best" possible solution. Rather, the GA is highly efficient in quickly yielding a "good" and "robust" solution rated against a user-defined fitness criterion. The GAabbreviate uses the GA package (Scrucca, 2013) to efficiently implement Yarkoni's (2010) scale abbreviation cost function: Cost = Ik + ∑_{i=1}^s w_i(1-R_i^2) where I represents a user-specified fixed item cost, k represents the number of items retained by the GA (in any given iteration), s is the number of subscales in the measure, w_i are the weights (by default w_i = 1 for any i) associated with each subscale (if there are any subsets to be retained), and R_i^2 is the amount of variance in the ith subscale that can be explained by a linear combination of individual item scores. Adjusting the value of I low or high yields longer or shorter measures respectively. When the cost of each individual item retained in each generation outweighs the cost of a loss in explained variance, the GA yields a relatively brief measure. When the cost is low, the GA yields a relatively longer measure maximizing explained variance (Yarkoni, 2010). Sahdra, Ciarrochi, Parker & Scrucca (2016) contains an example of how ValueAn object of class
A Author(s)Luca Scrucca, Department of Economics, University of Perugia, Perugia, ITALY Baljinder K. Sahdra, Institute for Positive Psychology and Education, Australian Catholic University, Strathfield, NSW, AUSTRALIA Send inquiries to baljinder.sahdra@acu.edu.au. ReferencesSahdra B. K., Ciarrochi J., Parker P. and Scrucca L. (2016). Using genetic algorithms in a large nationally representative American sample to abbreviate the Multidimensional Experiential Avoidance Questionnaire. Frontiers in Psychology, Volume 7(189), pp. 1–14. http://www.frontiersin.org/quantitative_psychology_and_measurement/10.3389/fpsyg.2016.00189/abstract Scrucca, L. (2013). GA: a package for genetic algorithms in R. Journal of Statistical Software, 53(4), 1-37, http://www.jstatsoft.org/v53/i04/. Yarkoni, T. (2010). The abbreviation of personality, or how to measure 200 personality scales with 200 items. Journal of Research in Personality, 44(2), 180-198. See Alsoga Examples
### Example using random generated data
nsubject = 100
nitems = 15
set.seed(123)
items = matrix(sample(1:5, nsubject*nitems, replace = TRUE),
nrow = nsubject, ncol = nitems)
scales = cbind(rowSums(items[,1:10]), rowSums(items[,11:15]))
GAA = GAabbreviate(items, scales, itemCost = 0.01, maxItems = 5,
popSize = 50, maxiter = 300, run = 100)
plot(GAA)
summary(GAA)
# more info can be retrieved using
GAA$best
GAA$measure
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(GAabbreviate)
Loading required package: GA
Loading required package: foreach
Loading required package: iterators
Package 'GA' version 3.0.2
Type 'citation("GA")' for citing this R package in publications.
Loading required package: psych
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/GAabbreviate/GAabbreviate.Rd_%03d_medium.png", width=480, height=480)
> ### Name: GAabbreviate
> ### Title: Abbreviating items (from questionnaire or other) measures using
> ### Genetic Algorithms (GAs)
> ### Aliases: GAabbreviate
> ### Keywords: optimize multivariate survey
>
> ### ** Examples
>
> ### Example using random generated data
> nsubject = 100
> nitems = 15
> set.seed(123)
> items = matrix(sample(1:5, nsubject*nitems, replace = TRUE),
+ nrow = nsubject, ncol = nitems)
> scales = cbind(rowSums(items[,1:10]), rowSums(items[,11:15]))
>
> GAA = GAabbreviate(items, scales, itemCost = 0.01, maxItems = 5,
+ popSize = 50, maxiter = 300, run = 100)
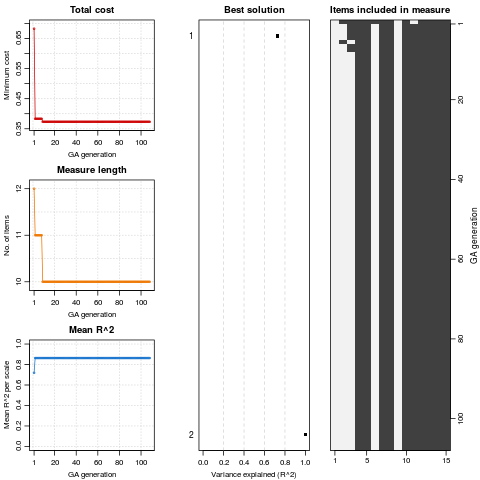
> plot(GAA)
> summary(GAA)
+-----------------------------------+
| Genetic Algorithm |
+-----------------------------------+
GA settings:
Type = binary
Population size = 50
Number of generations = 300
Elitism = 2
Crossover probability = 0.8
Mutation probability = 0.1
GA results:
Iterations = 108
Total cost = 0.3731952
Number of items in initial set = 15
Number of items in final set = 10
Mean coefficient alpha = 0.04562
Mean convergent correlation (training) = 0.9266
Mean convergent correlation (validation) = 0.9266
> # more info can be retrieved using
> GAA$best
$cost
[1] 0.3731952
$fit
[1] 0.2731952 0.0000000
> GAA$measure
$items
x4 x5 x7 x8 x10 x11 x12 x13 x14 x15
4 5 7 8 10 11 12 13 14 15
$nItems
[1] 10
$key
[,1] [,2]
[1,] 1 0
[2,] 1 0
[3,] 1 0
[4,] 1 0
[5,] 1 0
[6,] 0 1
[7,] 0 1
[8,] 0 1
[9,] 0 1
[10,] 0 1
$nScaleItems
[1] 5 5
$alpha
A1 A2
alpha 0.1355966 -0.04436475
$ccTraining
[1] 0.8532731 1.0000000
$ccValidation
[1] 0.8532101 1.0000000
>
>
>
>
>
> dev.off()
null device
1
>
|