Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Fit Generalized Semiparametric Mixed-Effects ModelsDescriptionFit a semiparametric mixed model or a generalized semiparametric mixed model. Usage
bGAMM(fix=formula, add=formula, rnd=formula,
data, lambda, family = NULL, control = list())
Arguments
ValueGeneric functions such as
Author(s)Andreas Groll andreas.groll@stat.uni-muenchen.de ReferencesGroll, A. and G. Tutz (2012). Regularization for Generalized Additive Mixed Models by Likelihood-Based Boosting. Methods of Information in Medicine 51(2), 168–177. See Also
Examples
data("soccer")
gamm1 <- bGAMM(points ~ ball.possession + tackles,
~ transfer.spendings + transfer.receits
+ unfair.score + ave.attend + sold.out,
rnd = list(team=~1), data = soccer, lambda = 1e+5,
family = poisson(link = log), control = list(steps=200, overdispersion=TRUE,
start=c(1,rep(0,25))))
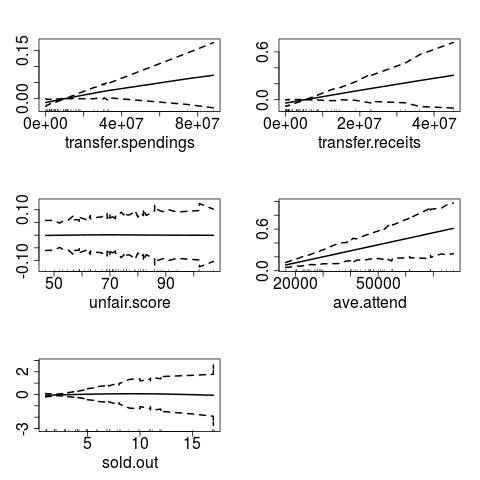
plot(gamm1)
# see also demo("bGAMM-soccer")
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(GMMBoost)
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/GMMBoost/bGAMM.rd_%03d_medium.png", width=480, height=480)
> ### Name: bGAMM
> ### Title: Fit Generalized Semiparametric Mixed-Effects Models
> ### Aliases: bGAMM
> ### Keywords: models methods
>
> ### ** Examples
>
> data("soccer")
>
> gamm1 <- bGAMM(points ~ ball.possession + tackles,
+ ~ transfer.spendings + transfer.receits
+ + unfair.score + ave.attend + sold.out,
+ rnd = list(team=~1), data = soccer, lambda = 1e+5,
+ family = poisson(link = log), control = list(steps=200, overdispersion=TRUE,
+ start=c(1,rep(0,25))))
[1] "Iteration 1"
[1] "Iteration 2"
[1] "Iteration 3"
[1] "Iteration 4"
[1] "Iteration 5"
[1] "Iteration 6"
[1] "Iteration 7"
[1] "Iteration 8"
[1] "Iteration 9"
[1] "Iteration 10"
[1] "Iteration 11"
[1] "Iteration 12"
[1] "Iteration 13"
[1] "Iteration 14"
[1] "Iteration 15"
[1] "Iteration 16"
[1] "Iteration 17"
[1] "Iteration 18"
[1] "Iteration 19"
[1] "Iteration 20"
[1] "Iteration 21"
[1] "Iteration 22"
[1] "Iteration 23"
[1] "Iteration 24"
[1] "Iteration 25"
[1] "Iteration 26"
[1] "Iteration 27"
[1] "Iteration 28"
[1] "Iteration 29"
[1] "Iteration 30"
[1] "Iteration 31"
[1] "Iteration 32"
[1] "Iteration 33"
[1] "Iteration 34"
[1] "Iteration 35"
[1] "Iteration 36"
[1] "Iteration 37"
[1] "Iteration 38"
[1] "Iteration 39"
[1] "Iteration 40"
[1] "Iteration 41"
[1] "Iteration 42"
[1] "Iteration 43"
[1] "Iteration 44"
[1] "Iteration 45"
[1] "Iteration 46"
[1] "Iteration 47"
[1] "Iteration 48"
[1] "Iteration 49"
[1] "Iteration 50"
[1] "Iteration 51"
[1] "Iteration 52"
[1] "Iteration 53"
[1] "Iteration 54"
[1] "Iteration 55"
[1] "Iteration 56"
[1] "Iteration 57"
[1] "Iteration 58"
[1] "Iteration 59"
[1] "Iteration 60"
[1] "Iteration 61"
[1] "Iteration 62"
[1] "Iteration 63"
[1] "Iteration 64"
[1] "Iteration 65"
[1] "Iteration 66"
[1] "Iteration 67"
[1] "Iteration 68"
[1] "Iteration 69"
[1] "Iteration 70"
[1] "Iteration 71"
[1] "Iteration 72"
[1] "Iteration 73"
[1] "Iteration 74"
[1] "Iteration 75"
[1] "Iteration 76"
[1] "Iteration 77"
[1] "Iteration 78"
[1] "Iteration 79"
[1] "Iteration 80"
[1] "Iteration 81"
[1] "Iteration 82"
[1] "Iteration 83"
[1] "Iteration 84"
[1] "Iteration 85"
[1] "Iteration 86"
[1] "Iteration 87"
[1] "Iteration 88"
[1] "Iteration 89"
[1] "Iteration 90"
[1] "Iteration 91"
[1] "Iteration 92"
[1] "Iteration 93"
[1] "Iteration 94"
[1] "Iteration 95"
[1] "Iteration 96"
[1] "Iteration 97"
[1] "Iteration 98"
[1] "Iteration 99"
[1] "Iteration 100"
[1] "Iteration 101"
[1] "Iteration 102"
[1] "Iteration 103"
[1] "Iteration 104"
[1] "Iteration 105"
[1] "Iteration 106"
[1] "Iteration 107"
[1] "Iteration 108"
[1] "Iteration 109"
[1] "Iteration 110"
[1] "Iteration 111"
[1] "Iteration 112"
[1] "Iteration 113"
[1] "Iteration 114"
[1] "Iteration 115"
[1] "Iteration 116"
[1] "Iteration 117"
[1] "Iteration 118"
[1] "Iteration 119"
[1] "Iteration 120"
[1] "Iteration 121"
[1] "Iteration 122"
[1] "Iteration 123"
[1] "Iteration 124"
[1] "Iteration 125"
[1] "Iteration 126"
[1] "Iteration 127"
[1] "Iteration 128"
[1] "Iteration 129"
[1] "Iteration 130"
[1] "Iteration 131"
[1] "Iteration 132"
[1] "Iteration 133"
[1] "Iteration 134"
[1] "Iteration 135"
[1] "Iteration 136"
[1] "Iteration 137"
[1] "Iteration 138"
[1] "Iteration 139"
[1] "Iteration 140"
[1] "Iteration 141"
[1] "Iteration 142"
[1] "Iteration 143"
[1] "Iteration 144"
[1] "Iteration 145"
[1] "Iteration 146"
[1] "Iteration 147"
[1] "Iteration 148"
[1] "Iteration 149"
[1] "Iteration 150"
[1] "Iteration 151"
[1] "Iteration 152"
[1] "Iteration 153"
[1] "Iteration 154"
[1] "Iteration 155"
[1] "Iteration 156"
[1] "Iteration 157"
[1] "Iteration 158"
[1] "Iteration 159"
[1] "Iteration 160"
[1] "Iteration 161"
[1] "Iteration 162"
[1] "Iteration 163"
[1] "Iteration 164"
[1] "Iteration 165"
[1] "Iteration 166"
[1] "Iteration 167"
[1] "Iteration 168"
[1] "Iteration 169"
[1] "Iteration 170"
[1] "Iteration 171"
[1] "Iteration 172"
[1] "Iteration 173"
[1] "Iteration 174"
[1] "Iteration 175"
[1] "Iteration 176"
[1] "Iteration 177"
[1] "Iteration 178"
[1] "Iteration 179"
[1] "Iteration 180"
[1] "Iteration 181"
[1] "Iteration 182"
[1] "Iteration 183"
[1] "Iteration 184"
[1] "Iteration 185"
[1] "Iteration 186"
[1] "Iteration 187"
[1] "Iteration 188"
[1] "Iteration 189"
[1] "Iteration 190"
[1] "Iteration 191"
[1] "Iteration 192"
[1] "Iteration 193"
[1] "Iteration 194"
[1] "Iteration 195"
[1] "Iteration 196"
[1] "Iteration 197"
[1] "Iteration 198"
[1] "Iteration 199"
[1] "Iteration 200"
>
> plot(gamm1)
>
> # see also demo("bGAMM-soccer")
>
>
>
>
>
> dev.off()
null device
1
>
|