Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Methods to fit a regression-kriging modelDescriptionTries to automatically fit a 2D or 3D regression-kriging model for a given set of points (object of type Usage
## S4 method for signature
## 'SpatialPointsDataFrame,formula,SpatialPixelsDataFrame'
fit.gstatModel(observations, formulaString, covariates,
method = list("GLM", "rpart", "randomForest", "quantregForest",
"xgboost", "ranger"),
dimensions = list("2D", "3D", "2D+T", "3D+T"),
fit.family = gaussian(), stepwise = TRUE, vgmFun = "Exp",
subsample = 5000, subsample.reg = 10000, ...)
## S4 method for signature 'geosamples,formula,SpatialPixelsDataFrame'
fit.gstatModel(observations, formulaString, covariates,
method = list("GLM", "rpart", "randomForest", "quantregForest",
"xgboost", "ranger"),
dimensions = list("2D", "3D", "2D+T", "3D+T"),
fit.family = gaussian(), stepwise = TRUE,
vgmFun = "Exp", subsample = 5000, subsample.reg = 10000, ...)
## S4 method for signature 'geosamples,formula,list'
fit.gstatModel(observations, formulaString, covariates,
method = list("GLM", "rpart", "randomForest", "quantregForest",
"xgboost", "ranger"),
dimensions = list("2D", "3D", "2D+T", "3D+T"),
fit.family = gaussian(), stepwise = TRUE,
vgmFun = "Exp", subsample = 5000, subsample.reg = 10000, ...)
## S4 method for signature 'geosamples,list,list'
fit.gstatModel(observations, formulaString, covariates,
method = list("GLM", "rpart", "randomForest", "quantregForest",
"xgboost", "ranger"),
dimensions = list("2D", "3D", "2D+T", "3D+T"),
fit.family = gaussian(), stepwise = TRUE,
vgmFun = "Exp", subsample = 5000, subsample.reg = 10000, ...)
Arguments
DetailsThe GLM method by default assumes that the target variable follows a normal distribution
NoteResiduals (response residuals from the model) will be checked for normality and problems reported by default. The warning messages should be taken with care, as when the sample size is small, even big departures from normality will not be reported; when the sample size is large, even the smallest deviation from normality might lead to a warning. Likewise, if the variogram fitting fails, consider fitting a variogram manually or using the Author(s)Tomislav Hengl, Gerard B.M. Heuvelink and Bas Kempen References
See Also
Examples
# 2D model:
library(sp)
library(boot)
library(aqp)
library(plyr)
library(rpart)
library(splines)
library(gstat)
library(randomForest)
library(quantregForest)
library(plotKML)
## load the Meuse data set:
demo(meuse, echo=FALSE)
## simple model:
omm <- fit.gstatModel(meuse, om~dist+ffreq, meuse.grid,
family = gaussian(log))
om.rk <- predict(omm, meuse.grid)
plot(om.rk)
## it was succesful!
## fit a GLM with a gaussian log-link:
omm <- fit.gstatModel(meuse, om~dist+ffreq, meuse.grid,
fit.family = gaussian(log))
summary(omm@regModel)
om.rk <- predict(omm, meuse.grid)
plot(om.rk)
## fit a regression-tree:
omm <- fit.gstatModel(meuse, log1p(om)~dist+ffreq, meuse.grid,
method="rpart")
summary(omm@regModel)
## plot a regression-tree:
plot(omm@regModel, uniform=TRUE)
text(omm@regModel, use.n=TRUE, all=TRUE, cex=.8)
omm@vgmModel
## fit a randomForest model:
omm <- fit.gstatModel(meuse, om~dist+ffreq, meuse.grid,
method="randomForest")
## plot to see how good is the fit:
plot(omm)
## plot the estimated error for number of bootstrapped trees:
plot(omm@regModel)
omm@vgmModel
om.rk <- predict(omm, meuse.grid)
plot(om.rk)
## Compare with "quantregForest" package:
omm <- fit.gstatModel(meuse, om~dist+ffreq, meuse.grid,
method="quantregForest")
## Not run:
om.rk <- predict(omm, meuse.grid, nfold=0)
plot(om.rk)
## plot the results in Google Earth:
plotKML(om.rk)
## End(Not run)
## binary variable (0/1):
meuse$soil.1 <- as.numeric(I(meuse$soil==1))
som <- fit.gstatModel(meuse, soil.1~dist+ffreq, meuse.grid,
fit.family = binomial(logit))
summary(som@regModel)
som.rk <- predict(som, meuse.grid)
plot(som.rk)
## Not run: # plot the results in Google Earth:
plotKML(som.rk)
## End(Not run)
## 3D model:
library(plotKML)
data(eberg)
## list columns of interest:
s.lst <- c("ID", "soiltype", "TAXGRSC", "X", "Y")
h.lst <- c("UHDICM","LHDICM","SNDMHT","SLTMHT","CLYMHT")
sel <- runif(nrow(eberg))<.05
## get sites table:
sites <- eberg[sel,s.lst]
## get horizons table:
horizons <- getHorizons(eberg[sel,], idcol="ID", sel=h.lst)
## create object of type "SoilProfileCollection"
eberg.spc <- join(horizons, sites, type='inner')
depths(eberg.spc) <- ID ~ UHDICM + LHDICM
site(eberg.spc) <- as.formula(paste("~", paste(s.lst[-1], collapse="+"), sep=""))
coordinates(eberg.spc) <- ~X+Y
proj4string(eberg.spc) <- CRS("+init=epsg:31467")
## convert to geosamples:
eberg.geo <- as.geosamples(eberg.spc)
## covariates:
data(eberg_grid)
gridded(eberg_grid) <- ~x+y
proj4string(eberg_grid) <- CRS("+init=epsg:31467")
glm.formulaString = as.formula(paste("SNDMHT ~ ",
paste(names(eberg_grid), collapse="+"), "+ ns(altitude, df=4)"))
SNDMHT.m <- fit.gstatModel(observations=eberg.geo, glm.formulaString,
covariates=eberg_grid)
plot(SNDMHT.m)
## problems with the variogram?
## Not run: ## remove classes from the PRMGEO6 that are not represented in the model:
sel = !(levels(eberg_grid$PRMGEO6) %in% levels(SNDMHT.m@regModel$model$PRMGEO6))
fix.c = levels(eberg_grid$PRMGEO6)[sel]
summary(eberg_grid$PRMGEO6)
for(j in fix.c){
eberg_grid$PRMGEO6[eberg_grid$PRMGEO6 == j] <- levels(eberg_grid$PRMGEO6)[7]
}
## prepare new locations:
new3D <- sp3D(eberg_grid)
## regression only:
SNDMHT.rk.sd1 <- predict(SNDMHT.m, new3D[[1]], vgmmodel=NULL)
## regression-kriging:
SNDMHT.rk.sd1 <- predict(SNDMHT.m, new3D[[1]])
## plot the results in Google Earth:
plotKML(SNDMHT.rk.sd1, z.lim=c(5,85))
## End(Not run)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(GSIF)
GSIF version 0.5-2 (2016-06-25)
URL: http://gsif.r-forge.r-project.org/
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/GSIF/fit.gstatModel.Rd_%03d_medium.png", width=480, height=480)
> ### Name: fit.gstatModel-methods
> ### Title: Methods to fit a regression-kriging model
> ### Aliases: fit.gstatModel-method fit.gstatModel
> ### fit.gstatModel,SpatialPointsDataFrame,formula,SpatialPixelsDataFrame-method
> ### fit.gstatModel,geosamples,formula,SpatialPixelsDataFrame-method
> ### fit.gstatModel,geosamples,formula,list-method
> ### fit.gstatModel,geosamples,list,list-method
> ### Keywords: methods
>
> ### ** Examples
>
> # 2D model:
> library(sp)
> library(boot)
> library(aqp)
This is aqp 1.9.3
> library(plyr)
> library(rpart)
> library(splines)
> library(gstat)
> library(randomForest)
randomForest 4.6-12
Type rfNews() to see new features/changes/bug fixes.
> library(quantregForest)
Loading required package: RColorBrewer
> library(plotKML)
plotKML version 0.5-6 (2016-05-02)
URL: http://plotkml.r-forge.r-project.org/
>
> ## load the Meuse data set:
> demo(meuse, echo=FALSE)
>
> ## simple model:
> omm <- fit.gstatModel(meuse, om~dist+ffreq, meuse.grid,
+ family = gaussian(log))
Fitting a linear model...
Fitting a 2D variogram...
Saving an object of class 'gstatModel'...
> om.rk <- predict(omm, meuse.grid)
Subsetting observations to fit the prediction domain in 2D...
Generating predictions using the trend model (RK method)...
[using ordinary kriging]
45% done 100% done
Running 5-fold cross validation using 'krige.cv'...
Creating an object of class "SpatialPredictions"
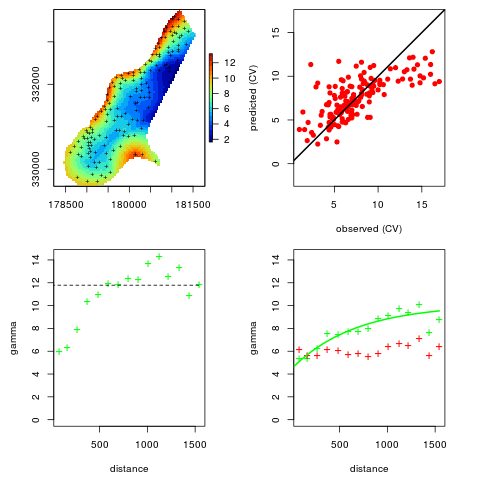
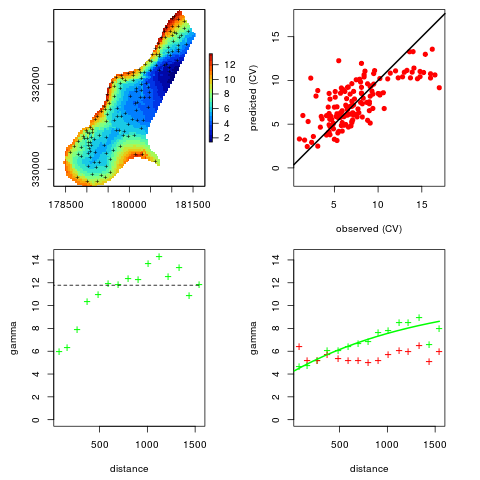
> plot(om.rk)
> ## it was succesful!
>
> ## fit a GLM with a gaussian log-link:
> omm <- fit.gstatModel(meuse, om~dist+ffreq, meuse.grid,
+ fit.family = gaussian(log))
Fitting a 2D variogram...
Saving an object of class 'gstatModel'...
> summary(omm@regModel)
Call:
glm(formula = om ~ dist + ffreq, family = fit.family, data = rmatrix)
Deviance Residuals:
Min 1Q Median 3Q Max
-7.5173 -1.9150 0.2131 1.7337 8.0391
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.42319 0.03958 61.215 < 2e-16 ***
dist -1.73313 0.20995 -8.255 7.49e-14 ***
ffreq2 -0.13713 0.06780 -2.023 0.0449 *
ffreq3 -0.11240 0.09236 -1.217 0.2255
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for gaussian family taken to be 6.988786)
Null deviance: 1791.4 on 152 degrees of freedom
Residual deviance: 1041.3 on 149 degrees of freedom
(2 observations deleted due to missingness)
AIC: 737.62
Number of Fisher Scoring iterations: 7
> om.rk <- predict(omm, meuse.grid)
Subsetting observations to fit the prediction domain in 2D...
Generating predictions using the trend model (RK method)...
[using ordinary kriging]
100% done
Running 5-fold cross validation using 'krige.cv'...
Creating an object of class "SpatialPredictions"
> plot(om.rk)
>
> ## fit a regression-tree:
> omm <- fit.gstatModel(meuse, log1p(om)~dist+ffreq, meuse.grid,
+ method="rpart")
Fitting a regression tree model...
Estimated Complexity Parameter (for prunning): 0.0371
Warning: Shapiro-Wilk normality test and Anderson-Darling normality test report probability of < .05 indicating lack of normal distribution for residuals
Fitting a 2D variogram...
Saving an object of class 'gstatModel'...
> summary(omm@regModel)
Call:
rpart::rpart(formula = formulaString, data = rmatrix.s)
n=153 (2 observations deleted due to missingness)
CP nsplit rel error xerror xstd
1 0.40702329 0 1.0000000 1.0171084 0.13444449
2 0.03709721 1 0.5929767 0.6245742 0.09430749
Variable importance
dist
100
Node number 1: 153 observations, complexity param=0.4070233
mean=2.05616, MSE=0.1702532
left son=2 (116 obs) right son=3 (37 obs)
Primary splits:
dist < 0.07302095 to the right, improve=0.40702330, (0 missing)
ffreq splits as RLL, improve=0.07840131, (0 missing)
Node number 2: 116 observations
mean=1.907488, MSE=0.1125991
Node number 3: 37 observations
mean=2.522267, MSE=0.06445407
> ## plot a regression-tree:
> plot(omm@regModel, uniform=TRUE)
> text(omm@regModel, use.n=TRUE, all=TRUE, cex=.8)
> omm@vgmModel
model psill range kappa ang1 ang2 ang3 anis1 anis2
1 Nug 3.684982 0.000 0.0 0 0 0 1 1
2 Exp 9.090300 498.053 0.5 0 0 0 1 1
>
> ## fit a randomForest model:
> omm <- fit.gstatModel(meuse, om~dist+ffreq, meuse.grid,
+ method="randomForest")
Fitting a randomForest model...
Fitting a 2D variogram...
Saving an object of class 'gstatModel'...
> ## plot to see how good is the fit:
> plot(omm)
Error in dev.new(width = 9, height = 5) :
no suitable unused file name for pdf()
Calls: plot -> plot -> dev.new
Execution halted
|