Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Generalized UniFrac distances for comparing microbial communities.DescriptionA generalized version of commonly used UniFrac distances. It is defined as: d^{(α)} = [∑_{i=1}^m b_i (p^A_{i} + p^B_{i})^α |p^A_{i} - p^B_{i}|/(p^A_{i} + p^B_{i})]/ [ ∑_{i=1}^m b_i (p^A_{i} + p^B_{i})^α], where m is the number of branches, b_i is the length of ith branch, p^A_{i}, p^B_{i} are the branch proportion for community A and B. Generalized UniFrac distance contains an extra parameter α controlling the weight on abundant lineages so the distance is not dominated by highly abundant lineages. α=0.5 has overall the best power. The unweighted and weighted UniFrac, and variance adjusted weighted UniFrac distances are also implemented. UsageGUniFrac(otu.tab, tree, alpha = c(0, 0.5, 1)) Arguments
ValueReturn a LIST containing
NoteThe function only accepts rooted tree. To root a tree, you may consider using the R code from https://stat.ethz.ch/pipermail/r-sig-phylo/2010-September/000750.html Author(s)Jun Chen <chenjun@mail.med.upenn.edu> ReferencesJun Chen and Hongzhe Li(2012). Associating microbiome composition with environmental covariates using generalized UniFrac distances. (Submitted) See Also
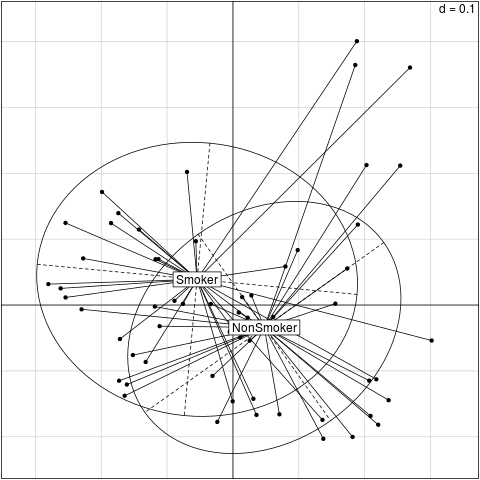
Examplesrequire(ade4) data(throat.otu.tab) data(throat.tree) data(throat.meta) groups <- throat.meta$SmokingStatus # Rarefaction otu.tab.rff <- Rarefy(throat.otu.tab)$otu.tab.rff # Calculate the UniFracs unifracs <- GUniFrac(otu.tab.rff, throat.tree, alpha=c(0, 0.5, 1))$unifracs dw <- unifracs[, , "d_1"] # Weighted UniFrac du <- unifracs[, , "d_UW"] # Unweighted UniFrac dv <- unifracs[, , "d_VAW"] # Variance adjusted weighted UniFrac d0 <- unifracs[, , "d_0"] # GUniFrac with alpha 0 d5 <- unifracs[, , "d_0.5"] # GUniFrac with alpha 0.5 # Permanova - Distance based multivariate analysis of variance adonis(as.dist(d5) ~ groups) # PCoA plot s.class(cmdscale(d5, k=2), fac = groups) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(GUniFrac)
Loading required package: vegan
Loading required package: permute
Loading required package: lattice
This is vegan 2.4-0
Loading required package: ape
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/GUniFrac/GUniFrac.Rd_%03d_medium.png", width=480, height=480)
> ### Name: GUniFrac
> ### Title: Generalized UniFrac distances for comparing microbial
> ### communities.
> ### Aliases: GUniFrac
> ### Keywords: distance UniFrac ecology
>
> ### ** Examples
>
> require(ade4)
Loading required package: ade4
Attaching package: 'ade4'
The following object is masked from 'package:vegan':
cca
>
> data(throat.otu.tab)
> data(throat.tree)
> data(throat.meta)
>
> groups <- throat.meta$SmokingStatus
>
> # Rarefaction
> otu.tab.rff <- Rarefy(throat.otu.tab)$otu.tab.rff
>
> # Calculate the UniFracs
> unifracs <- GUniFrac(otu.tab.rff, throat.tree, alpha=c(0, 0.5, 1))$unifracs
>
> dw <- unifracs[, , "d_1"] # Weighted UniFrac
> du <- unifracs[, , "d_UW"] # Unweighted UniFrac
> dv <- unifracs[, , "d_VAW"] # Variance adjusted weighted UniFrac
> d0 <- unifracs[, , "d_0"] # GUniFrac with alpha 0
> d5 <- unifracs[, , "d_0.5"] # GUniFrac with alpha 0.5
>
> # Permanova - Distance based multivariate analysis of variance
> adonis(as.dist(d5) ~ groups)
Call:
adonis(formula = as.dist(d5) ~ groups)
Permutation: free
Number of permutations: 999
Terms added sequentially (first to last)
Df SumsOfSqs MeanSqs F.Model R2 Pr(>F)
groups 1 0.3500 0.35004 2.501 0.04134 0.002 **
Residuals 58 8.1179 0.13996 0.95866
Total 59 8.4680 1.00000
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
>
> # PCoA plot
> s.class(cmdscale(d5, k=2), fac = groups)
>
>
>
>
>
>
> dev.off()
null device
1
>
|