Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Graphical Gaussian Models: Assess Significance of Edges (and Directions)Description
Usage
network.test.edges(r.mat, fdr=TRUE, direct=FALSE, plot=TRUE, ...)
extract.network(network.all, method.ggm=c("prob", "qval","number"),
cutoff.ggm=0.8, method.dir=c("prob","qval","number", "all"),
cutoff.dir=0.8, verbose=TRUE)
Arguments
DetailsFor assessing the significance of edges in the GGM
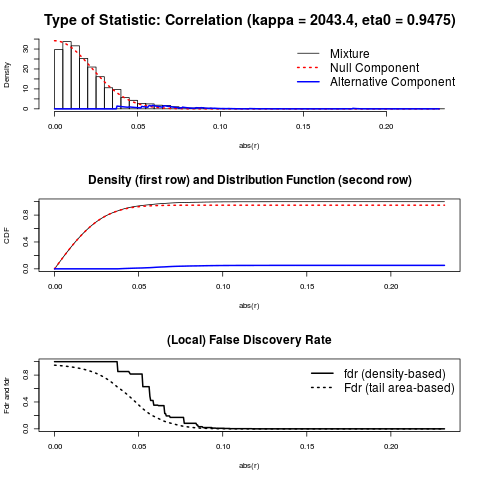
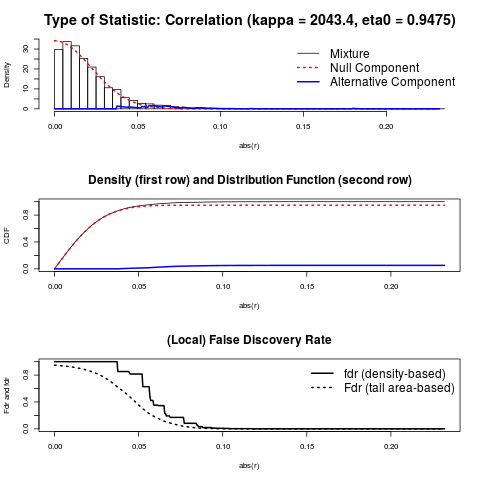
a mixture model is fitted to the partial correlations using For determining putatative directions on this GGM log-ratios of standardized partial variances re estimated, and subsequently the corresponding (local) fdr values are computed - see Opgen-Rhein and Strimmer (2007). Value
Each row in the data frame corresponds to one edge, and the rows are sorted according the absolute strength of the correlation (from strongest to weakest)
Author(s)Rainer Opgen-Rhein, Juliane Sch"afer, Korbinian Strimmer (http://strimmerlab.org). ReferencesSch"afer, J., and Strimmer, K. (2005). An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics 21:754-764. Opgen-Rhein, R., and K. Strimmer. (2007). From correlation to causation networks: a simple approximate learning algorithm and its application to high-dimensional plant gene expression data. BMC Syst. Biol. 1:37. See Also
Examples
# load GeneNet library
library("GeneNet")
# ecoli data
data(ecoli)
# estimate partial correlation matrix
inferred.pcor <- ggm.estimate.pcor(ecoli)
# p-values, q-values and posterior probabilities for each potential edge
#
test.results <- network.test.edges(inferred.pcor)
# show best 20 edges (strongest correlation)
test.results[1:20,]
# extract network containing edges with prob > 0.9 (i.e. local fdr < 0.1)
net <- extract.network(test.results, cutoff.ggm=0.9)
net
# how many are significant based on FDR cutoff Q=0.05 ?
num.significant.1 <- sum(test.results$qval <= 0.05)
test.results[1:num.significant.1,]
# how many are significant based on "local fdr" cutoff (prob > 0.9) ?
num.significant.2 <- sum(test.results$prob > 0.9)
test.results[test.results$prob > 0.9,]
# parameters of the mixture distribution used to compute p-values etc.
c <- fdrtool(sm2vec(inferred.pcor), statistic="correlation")
c$param
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(GeneNet)
Loading required package: corpcor
Loading required package: longitudinal
Loading required package: fdrtool
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/GeneNet/network.test.edges.Rd_%03d_medium.png", width=480, height=480)
> ### Name: network.test.edges
> ### Title: Graphical Gaussian Models: Assess Significance of Edges (and
> ### Directions)
> ### Aliases: network.test.edges extract.network
> ### Keywords: htest
>
> ### ** Examples
>
> # load GeneNet library
> library("GeneNet")
>
> # ecoli data
> data(ecoli)
>
> # estimate partial correlation matrix
> inferred.pcor <- ggm.estimate.pcor(ecoli)
Estimating optimal shrinkage intensity lambda (correlation matrix): 0.1804
>
> # p-values, q-values and posterior probabilities for each potential edge
> #
> test.results <- network.test.edges(inferred.pcor)
Estimate (local) false discovery rates (partial correlations):
Step 1... determine cutoff point
Step 2... estimate parameters of null distribution and eta0
Step 3... compute p-values and estimate empirical PDF/CDF
Step 4... compute q-values and local fdr
Step 5... prepare for plotting
>
> # show best 20 edges (strongest correlation)
> test.results[1:20,]
pcor node1 node2 pval qval prob
1 0.23185664 51 53 2.220446e-16 3.612205e-13 1.0000000
2 0.22405545 52 53 2.220446e-16 3.612205e-13 1.0000000
3 0.21507824 51 52 2.220446e-16 3.612205e-13 1.0000000
4 0.17328863 7 93 3.108624e-15 3.792816e-12 0.9999945
5 -0.13418892 29 86 1.120813e-09 1.093998e-06 0.9999516
6 0.12594697 21 72 1.103837e-08 8.978569e-06 0.9998400
7 0.11956105 28 86 5.890927e-08 3.853592e-05 0.9998400
8 -0.11723897 26 80 1.060526e-07 5.816175e-05 0.9998400
9 -0.11711625 72 89 1.093655e-07 5.930502e-05 0.9972804
10 0.10658013 20 21 1.366611e-06 5.925278e-04 0.9972804
11 0.10589778 21 73 1.596860e-06 6.678431e-04 0.9972804
12 0.10478689 20 91 2.053404e-06 8.024428e-04 0.9972804
13 0.10420836 7 52 2.338383e-06 8.778608e-04 0.9944557
14 0.10236077 87 95 3.525188e-06 1.224964e-03 0.9944557
15 0.10113550 27 95 4.610445e-06 1.500048e-03 0.9920084
16 0.09928954 21 51 6.868360e-06 2.046550e-03 0.9920084
17 0.09791914 21 88 9.192376e-06 2.520617e-03 0.9920084
18 0.09719685 18 95 1.070233e-05 2.790103e-03 0.9920084
19 0.09621791 28 90 1.313008e-05 3.171818e-03 0.9920084
20 0.09619099 12 80 1.320374e-05 3.182527e-03 0.9920084
>
> # extract network containing edges with prob > 0.9 (i.e. local fdr < 0.1)
> net <- extract.network(test.results, cutoff.ggm=0.9)
Significant edges: 65
Corresponding to 1.26 % of possible edges
> net
pcor node1 node2 pval qval prob
1 0.23185664 51 53 2.220446e-16 3.612205e-13 1.0000000
2 0.22405545 52 53 2.220446e-16 3.612205e-13 1.0000000
3 0.21507824 51 52 2.220446e-16 3.612205e-13 1.0000000
4 0.17328863 7 93 3.108624e-15 3.792816e-12 0.9999945
5 -0.13418892 29 86 1.120813e-09 1.093998e-06 0.9999516
6 0.12594697 21 72 1.103837e-08 8.978569e-06 0.9998400
7 0.11956105 28 86 5.890927e-08 3.853592e-05 0.9998400
8 -0.11723897 26 80 1.060526e-07 5.816175e-05 0.9998400
9 -0.11711625 72 89 1.093655e-07 5.930502e-05 0.9972804
10 0.10658013 20 21 1.366611e-06 5.925278e-04 0.9972804
11 0.10589778 21 73 1.596860e-06 6.678431e-04 0.9972804
12 0.10478689 20 91 2.053404e-06 8.024428e-04 0.9972804
13 0.10420836 7 52 2.338383e-06 8.778608e-04 0.9944557
14 0.10236077 87 95 3.525188e-06 1.224964e-03 0.9944557
15 0.10113550 27 95 4.610445e-06 1.500048e-03 0.9920084
16 0.09928954 21 51 6.868360e-06 2.046550e-03 0.9920084
17 0.09791914 21 88 9.192376e-06 2.520617e-03 0.9920084
18 0.09719685 18 95 1.070233e-05 2.790103e-03 0.9920084
19 0.09621791 28 90 1.313008e-05 3.171818e-03 0.9920084
20 0.09619099 12 80 1.320374e-05 3.182527e-03 0.9920084
21 0.09576091 89 95 1.443542e-05 3.354778e-03 0.9891317
22 0.09473210 7 51 1.784127e-05 3.864827e-03 0.9891317
23 -0.09386896 53 58 2.127623e-05 4.313591e-03 0.9891317
24 -0.09366615 29 83 2.217013e-05 4.421101e-03 0.9891317
25 -0.09341148 21 89 2.334321e-05 4.556948e-03 0.9810727
26 -0.09156391 49 93 3.380044e-05 5.955974e-03 0.9810727
27 -0.09150710 80 90 3.418364e-05 6.002084e-03 0.9810727
28 0.09101505 7 53 3.767967e-05 6.408104e-03 0.9810727
29 0.09050688 21 84 4.164472e-05 6.838785e-03 0.9810727
30 0.08965490 72 73 4.919367e-05 7.581868e-03 0.9810727
31 -0.08934025 29 99 5.229606e-05 7.861419e-03 0.9810727
32 -0.08906819 9 95 5.512710e-05 8.104761e-03 0.9810727
33 0.08888345 2 49 5.713146e-05 8.270675e-03 0.9810727
34 0.08850681 86 90 6.143364e-05 8.610164e-03 0.9810727
35 0.08805868 17 53 6.695172e-05 9.015178e-03 0.9810727
36 0.08790809 28 48 6.890886e-05 9.151294e-03 0.9810727
37 0.08783471 33 58 6.988213e-05 9.217600e-03 0.9682377
38 -0.08705796 7 49 8.101246e-05 1.021362e-02 0.9682377
39 0.08645033 20 46 9.086550e-05 1.102467e-02 0.9682377
40 0.08609950 48 86 9.705865e-05 1.150393e-02 0.9682377
41 0.08598769 21 52 9.911461e-05 1.165817e-02 0.9682377
42 0.08555275 32 95 1.075099e-04 1.226435e-02 0.9682377
43 0.08548231 17 51 1.089311e-04 1.236337e-02 0.9424721
44 0.08470370 80 83 1.258659e-04 1.382357e-02 0.9424721
45 0.08442510 80 82 1.325063e-04 1.437068e-02 0.9174572
46 0.08271606 80 93 1.810275e-04 1.845632e-02 0.9174572
47 0.08235175 46 91 1.933329e-04 1.941580e-02 0.9174572
48 0.08217787 25 95 1.994789e-04 1.988433e-02 0.9174572
49 -0.08170331 29 87 2.171999e-04 2.119715e-02 0.9174572
50 0.08123632 19 29 2.360717e-04 2.253606e-02 0.9174572
51 0.08101702 51 84 2.454547e-04 2.318025e-02 0.9174572
52 0.08030748 16 93 2.782643e-04 2.532796e-02 0.9174572
53 0.08006503 28 52 2.903870e-04 2.608272e-02 0.9174572
54 -0.07941656 41 80 3.252834e-04 2.814825e-02 0.9174572
55 0.07941410 54 89 3.254230e-04 2.815621e-02 0.9174572
56 -0.07934653 28 80 3.292785e-04 2.837512e-02 0.9174572
57 0.07916783 29 92 3.396803e-04 2.895702e-02 0.9174572
58 -0.07866905 17 86 3.703636e-04 3.060294e-02 0.9174572
59 0.07827749 16 29 3.962447e-04 3.191463e-02 0.9174572
60 -0.07808262 73 89 4.097453e-04 3.257290e-02 0.9174572
61 0.07766261 52 67 4.403166e-04 3.400207e-02 0.9174572
62 0.07762917 25 87 4.428397e-04 3.411638e-02 0.9174572
63 -0.07739378 9 93 4.609873e-04 3.492296e-02 0.9174572
64 0.07738885 31 80 4.613748e-04 3.493988e-02 0.9174572
65 -0.07718681 80 94 4.775137e-04 3.563445e-02 0.9174572
>
> # how many are significant based on FDR cutoff Q=0.05 ?
> num.significant.1 <- sum(test.results$qval <= 0.05)
> test.results[1:num.significant.1,]
pcor node1 node2 pval qval prob
1 0.23185664 51 53 2.220446e-16 3.612205e-13 1.0000000
2 0.22405545 52 53 2.220446e-16 3.612205e-13 1.0000000
3 0.21507824 51 52 2.220446e-16 3.612205e-13 1.0000000
4 0.17328863 7 93 3.108624e-15 3.792816e-12 0.9999945
5 -0.13418892 29 86 1.120813e-09 1.093998e-06 0.9999516
6 0.12594697 21 72 1.103837e-08 8.978569e-06 0.9998400
7 0.11956105 28 86 5.890927e-08 3.853592e-05 0.9998400
8 -0.11723897 26 80 1.060526e-07 5.816175e-05 0.9998400
9 -0.11711625 72 89 1.093655e-07 5.930502e-05 0.9972804
10 0.10658013 20 21 1.366611e-06 5.925278e-04 0.9972804
11 0.10589778 21 73 1.596860e-06 6.678431e-04 0.9972804
12 0.10478689 20 91 2.053404e-06 8.024428e-04 0.9972804
13 0.10420836 7 52 2.338383e-06 8.778608e-04 0.9944557
14 0.10236077 87 95 3.525188e-06 1.224964e-03 0.9944557
15 0.10113550 27 95 4.610445e-06 1.500048e-03 0.9920084
16 0.09928954 21 51 6.868360e-06 2.046550e-03 0.9920084
17 0.09791914 21 88 9.192376e-06 2.520617e-03 0.9920084
18 0.09719685 18 95 1.070233e-05 2.790103e-03 0.9920084
19 0.09621791 28 90 1.313008e-05 3.171818e-03 0.9920084
20 0.09619099 12 80 1.320374e-05 3.182527e-03 0.9920084
21 0.09576091 89 95 1.443542e-05 3.354778e-03 0.9891317
22 0.09473210 7 51 1.784127e-05 3.864827e-03 0.9891317
23 -0.09386896 53 58 2.127623e-05 4.313591e-03 0.9891317
24 -0.09366615 29 83 2.217013e-05 4.421101e-03 0.9891317
25 -0.09341148 21 89 2.334321e-05 4.556948e-03 0.9810727
26 -0.09156391 49 93 3.380044e-05 5.955974e-03 0.9810727
27 -0.09150710 80 90 3.418364e-05 6.002084e-03 0.9810727
28 0.09101505 7 53 3.767967e-05 6.408104e-03 0.9810727
29 0.09050688 21 84 4.164472e-05 6.838785e-03 0.9810727
30 0.08965490 72 73 4.919367e-05 7.581868e-03 0.9810727
31 -0.08934025 29 99 5.229606e-05 7.861419e-03 0.9810727
32 -0.08906819 9 95 5.512710e-05 8.104761e-03 0.9810727
33 0.08888345 2 49 5.713146e-05 8.270675e-03 0.9810727
34 0.08850681 86 90 6.143364e-05 8.610164e-03 0.9810727
35 0.08805868 17 53 6.695172e-05 9.015178e-03 0.9810727
36 0.08790809 28 48 6.890886e-05 9.151294e-03 0.9810727
37 0.08783471 33 58 6.988213e-05 9.217600e-03 0.9682377
38 -0.08705796 7 49 8.101246e-05 1.021362e-02 0.9682377
39 0.08645033 20 46 9.086550e-05 1.102467e-02 0.9682377
40 0.08609950 48 86 9.705865e-05 1.150393e-02 0.9682377
41 0.08598769 21 52 9.911461e-05 1.165817e-02 0.9682377
42 0.08555275 32 95 1.075099e-04 1.226435e-02 0.9682377
43 0.08548231 17 51 1.089311e-04 1.236337e-02 0.9424721
44 0.08470370 80 83 1.258659e-04 1.382357e-02 0.9424721
45 0.08442510 80 82 1.325063e-04 1.437068e-02 0.9174572
46 0.08271606 80 93 1.810275e-04 1.845632e-02 0.9174572
47 0.08235175 46 91 1.933329e-04 1.941580e-02 0.9174572
48 0.08217787 25 95 1.994789e-04 1.988433e-02 0.9174572
49 -0.08170331 29 87 2.171999e-04 2.119715e-02 0.9174572
50 0.08123632 19 29 2.360717e-04 2.253606e-02 0.9174572
51 0.08101702 51 84 2.454547e-04 2.318025e-02 0.9174572
52 0.08030748 16 93 2.782643e-04 2.532796e-02 0.9174572
53 0.08006503 28 52 2.903870e-04 2.608272e-02 0.9174572
54 -0.07941656 41 80 3.252834e-04 2.814825e-02 0.9174572
55 0.07941410 54 89 3.254230e-04 2.815621e-02 0.9174572
56 -0.07934653 28 80 3.292785e-04 2.837512e-02 0.9174572
57 0.07916783 29 92 3.396803e-04 2.895702e-02 0.9174572
58 -0.07866905 17 86 3.703636e-04 3.060294e-02 0.9174572
59 0.07827749 16 29 3.962447e-04 3.191463e-02 0.9174572
60 -0.07808262 73 89 4.097453e-04 3.257290e-02 0.9174572
61 0.07766261 52 67 4.403166e-04 3.400207e-02 0.9174572
62 0.07762917 25 87 4.428397e-04 3.411638e-02 0.9174572
63 -0.07739378 9 93 4.609873e-04 3.492296e-02 0.9174572
64 0.07738885 31 80 4.613748e-04 3.493988e-02 0.9174572
65 -0.07718681 80 94 4.775137e-04 3.563445e-02 0.9174572
66 0.07706275 27 58 4.876832e-04 3.606179e-02 0.8297810
67 -0.07610709 16 83 5.730534e-04 4.085920e-02 0.8297810
68 0.07550557 53 84 6.337144e-04 4.406473e-02 0.8297810
>
> # how many are significant based on "local fdr" cutoff (prob > 0.9) ?
> num.significant.2 <- sum(test.results$prob > 0.9)
> test.results[test.results$prob > 0.9,]
pcor node1 node2 pval qval prob
1 0.23185664 51 53 2.220446e-16 3.612205e-13 1.0000000
2 0.22405545 52 53 2.220446e-16 3.612205e-13 1.0000000
3 0.21507824 51 52 2.220446e-16 3.612205e-13 1.0000000
4 0.17328863 7 93 3.108624e-15 3.792816e-12 0.9999945
5 -0.13418892 29 86 1.120813e-09 1.093998e-06 0.9999516
6 0.12594697 21 72 1.103837e-08 8.978569e-06 0.9998400
7 0.11956105 28 86 5.890927e-08 3.853592e-05 0.9998400
8 -0.11723897 26 80 1.060526e-07 5.816175e-05 0.9998400
9 -0.11711625 72 89 1.093655e-07 5.930502e-05 0.9972804
10 0.10658013 20 21 1.366611e-06 5.925278e-04 0.9972804
11 0.10589778 21 73 1.596860e-06 6.678431e-04 0.9972804
12 0.10478689 20 91 2.053404e-06 8.024428e-04 0.9972804
13 0.10420836 7 52 2.338383e-06 8.778608e-04 0.9944557
14 0.10236077 87 95 3.525188e-06 1.224964e-03 0.9944557
15 0.10113550 27 95 4.610445e-06 1.500048e-03 0.9920084
16 0.09928954 21 51 6.868360e-06 2.046550e-03 0.9920084
17 0.09791914 21 88 9.192376e-06 2.520617e-03 0.9920084
18 0.09719685 18 95 1.070233e-05 2.790103e-03 0.9920084
19 0.09621791 28 90 1.313008e-05 3.171818e-03 0.9920084
20 0.09619099 12 80 1.320374e-05 3.182527e-03 0.9920084
21 0.09576091 89 95 1.443542e-05 3.354778e-03 0.9891317
22 0.09473210 7 51 1.784127e-05 3.864827e-03 0.9891317
23 -0.09386896 53 58 2.127623e-05 4.313591e-03 0.9891317
24 -0.09366615 29 83 2.217013e-05 4.421101e-03 0.9891317
25 -0.09341148 21 89 2.334321e-05 4.556948e-03 0.9810727
26 -0.09156391 49 93 3.380044e-05 5.955974e-03 0.9810727
27 -0.09150710 80 90 3.418364e-05 6.002084e-03 0.9810727
28 0.09101505 7 53 3.767967e-05 6.408104e-03 0.9810727
29 0.09050688 21 84 4.164472e-05 6.838785e-03 0.9810727
30 0.08965490 72 73 4.919367e-05 7.581868e-03 0.9810727
31 -0.08934025 29 99 5.229606e-05 7.861419e-03 0.9810727
32 -0.08906819 9 95 5.512710e-05 8.104761e-03 0.9810727
33 0.08888345 2 49 5.713146e-05 8.270675e-03 0.9810727
34 0.08850681 86 90 6.143364e-05 8.610164e-03 0.9810727
35 0.08805868 17 53 6.695172e-05 9.015178e-03 0.9810727
36 0.08790809 28 48 6.890886e-05 9.151294e-03 0.9810727
37 0.08783471 33 58 6.988213e-05 9.217600e-03 0.9682377
38 -0.08705796 7 49 8.101246e-05 1.021362e-02 0.9682377
39 0.08645033 20 46 9.086550e-05 1.102467e-02 0.9682377
40 0.08609950 48 86 9.705865e-05 1.150393e-02 0.9682377
41 0.08598769 21 52 9.911461e-05 1.165817e-02 0.9682377
42 0.08555275 32 95 1.075099e-04 1.226435e-02 0.9682377
43 0.08548231 17 51 1.089311e-04 1.236337e-02 0.9424721
44 0.08470370 80 83 1.258659e-04 1.382357e-02 0.9424721
45 0.08442510 80 82 1.325063e-04 1.437068e-02 0.9174572
46 0.08271606 80 93 1.810275e-04 1.845632e-02 0.9174572
47 0.08235175 46 91 1.933329e-04 1.941580e-02 0.9174572
48 0.08217787 25 95 1.994789e-04 1.988433e-02 0.9174572
49 -0.08170331 29 87 2.171999e-04 2.119715e-02 0.9174572
50 0.08123632 19 29 2.360717e-04 2.253606e-02 0.9174572
51 0.08101702 51 84 2.454547e-04 2.318025e-02 0.9174572
52 0.08030748 16 93 2.782643e-04 2.532796e-02 0.9174572
53 0.08006503 28 52 2.903870e-04 2.608272e-02 0.9174572
54 -0.07941656 41 80 3.252834e-04 2.814825e-02 0.9174572
55 0.07941410 54 89 3.254230e-04 2.815621e-02 0.9174572
56 -0.07934653 28 80 3.292785e-04 2.837512e-02 0.9174572
57 0.07916783 29 92 3.396803e-04 2.895702e-02 0.9174572
58 -0.07866905 17 86 3.703636e-04 3.060294e-02 0.9174572
59 0.07827749 16 29 3.962447e-04 3.191463e-02 0.9174572
60 -0.07808262 73 89 4.097453e-04 3.257290e-02 0.9174572
61 0.07766261 52 67 4.403166e-04 3.400207e-02 0.9174572
62 0.07762917 25 87 4.428397e-04 3.411638e-02 0.9174572
63 -0.07739378 9 93 4.609873e-04 3.492296e-02 0.9174572
64 0.07738885 31 80 4.613748e-04 3.493988e-02 0.9174572
65 -0.07718681 80 94 4.775137e-04 3.563445e-02 0.9174572
>
> # parameters of the mixture distribution used to compute p-values etc.
> c <- fdrtool(sm2vec(inferred.pcor), statistic="correlation")
Step 1... determine cutoff point
Step 2... estimate parameters of null distribution and eta0
Step 3... compute p-values and estimate empirical PDF/CDF
Step 4... compute q-values and local fdr
Step 5... prepare for plotting
> c$param
cutoff N.cens eta0 eta0.SE kappa kappa.SE
[1,] 0.03553068 4352 0.9474623 0.005656465 2043.377 94.72264
>
>
>
>
>
>
> dev.off()
null device
1
>
|