Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Fit the Hyperbolic Distribution to DataDescriptionFits a hyperbolic distribution to data. Displays the histogram, log-histogram (both with fitted densities), Q-Q plot and P-P plot for the fit which has the maximum likelihood. Usage
hyperbFit(x, freq = NULL, paramStart = NULL,
startMethod = c("Nelder-Mead","BFGS"),
startValues = c("BN","US","FN","SL","MoM"),
criterion = "MLE",
method = c("Nelder-Mead","BFGS","nlm",
"L-BFGS-B","nlminb","constrOptim"),
plots = FALSE, printOut = FALSE,
controlBFGS = list(maxit = 200),
controlNM = list(maxit = 1000), maxitNLM = 1500,
controlLBFGSB = list(maxit = 200),
controlNLMINB = list(),
controlCO = list(), ...)
## S3 method for class 'hyperbFit'
print(x,
digits = max(3, getOption("digits") - 3), ...)
## S3 method for class 'hyperbFit'
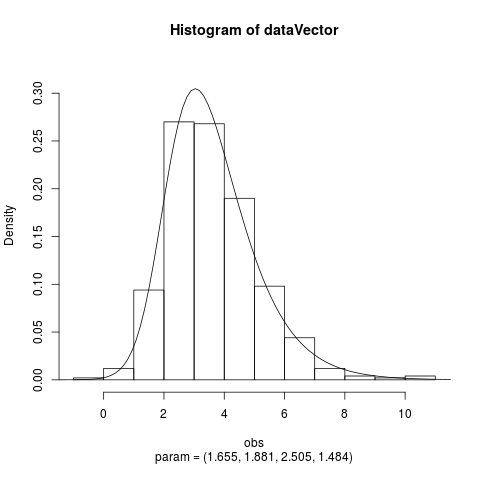
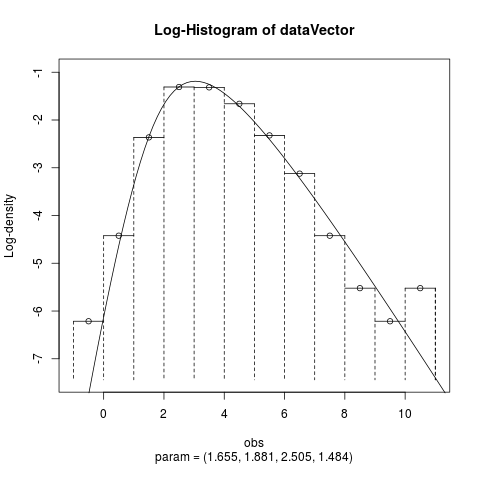
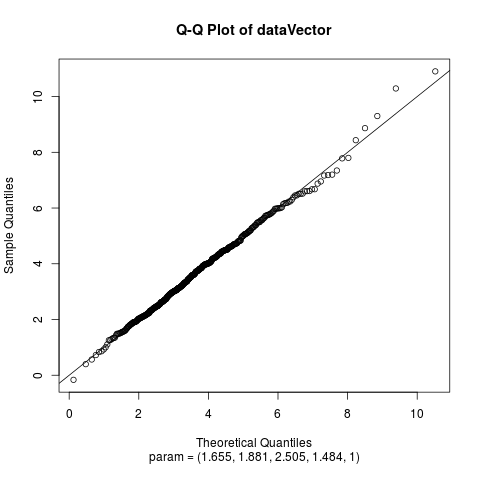
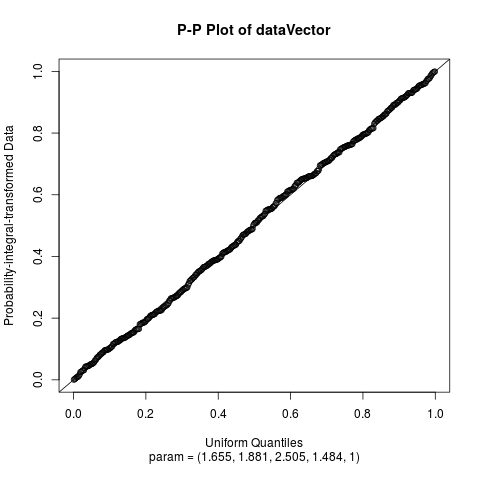
plot(x, which = 1:4,
plotTitles = paste(c("Histogram of ","Log-Histogram of ",
"Q-Q Plot of ","P-P Plot of "), x$obsName,
sep = ""),
ask = prod(par("mfcol")) < length(which) & dev.interactive(), ...)
## S3 method for class 'hyperbFit'
coef(object, ...)
## S3 method for class 'hyperbFit'
vcov(object, ...)
Arguments
Details
For the details concerning the use of The six optimisation methods currently available are:
For details of how to pass control information for optimisation using
When Value
Author(s)David Scott d.scott@auckland.ac.nz, Ai-Wei Lee, Jennifer Tso, Richard Trendall, Thomas Tran, Christine Yang Dong ReferencesBarndorff-Nielsen, O. (1977) Exponentially decreasing distributions for the logarithm of particle size, Proc. Roy. Soc. Lond., A353, 401–419. Fieller, N. J., Flenley, E. C. and Olbricht, W. (1992) Statistics of particle size data. Appl. Statist., 41, 127–146. See Also
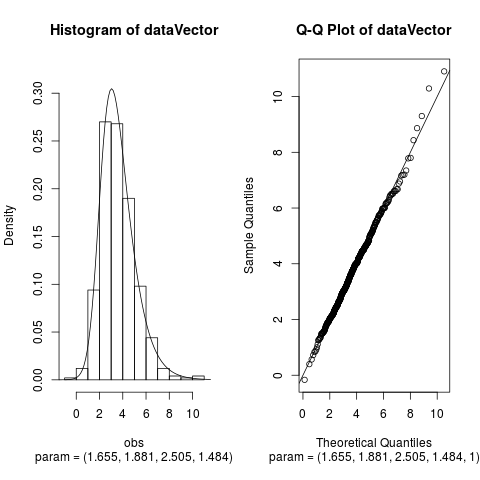
Examplesparam <- c(2, 2, 2, 1) dataVector <- rhyperb(500, param = param) ## See how well hyperbFit works hyperbFit(dataVector) hyperbFit(dataVector, plots = TRUE) fit <- hyperbFit(dataVector) par(mfrow = c(1, 2)) plot(fit, which = c(1, 3)) ## Use nlm instead of default hyperbFit(dataVector, method = "nlm") Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(GeneralizedHyperbolic)
Loading required package: DistributionUtils
Loading required package: RUnit
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/GeneralizedHyperbolic/hyperbFit.Rd_%03d_medium.png", width=480, height=480)
> ### Name: hyperbFit
> ### Title: Fit the Hyperbolic Distribution to Data
> ### Aliases: hyperbFit print.hyperbFit plot.hyperbFit coef.hyperbFit
> ### vcov.hyperbFit
> ### Keywords: distribution
>
> ### ** Examples
>
> param <- c(2, 2, 2, 1)
> dataVector <- rhyperb(500, param = param)
> ## See how well hyperbFit works
> hyperbFit(dataVector)
Data: dataVector
Parameter estimates:
mu delta alpha beta
1.655 1.881 2.505 1.484
Likelihood: -883.7581
criterion : MLE
Method: Nelder-Mead
Convergence code: 0
Iterations: 237
> hyperbFit(dataVector, plots = TRUE)
Data: dataVector
Parameter estimates:
mu delta alpha beta
1.655 1.881 2.505 1.484
Likelihood: -883.7581
criterion : MLE
Method: Nelder-Mead
Convergence code: 0
Iterations: 237
> fit <- hyperbFit(dataVector)
> par(mfrow = c(1, 2))
> plot(fit, which = c(1, 3))
>
> ## Use nlm instead of default
> hyperbFit(dataVector, method = "nlm")
Data: dataVector
Parameter estimates:
mu delta alpha beta
1.655 1.881 2.503 1.483
Likelihood: -883.758
criterion : MLE
Method: nlm
Convergence code: 1
Iterations: 27
Warning message:
In nlm(llfunc, paramStart, iterlim = maxitNLM, ...) :
NA/Inf replaced by maximum positive value
>
>
>
>
>
>
> dev.off()
null device
1
>
|