Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Estimation of Hierarchical Archimedean CopulaeDescriptionThe function estimates the parameters and determines the structure of Hierarchical Archimedean Copulae. Usageestimate.copula(X, type = 1, method = 1, hac = NULL, epsilon = 0, agg.method = "mean", margins = NULL, na.rm = FALSE, max.min = TRUE, ...) Arguments
ValueA hac object is returned. ReferencesGenest, C., Ghoudi, K., and Rivest, L. P. 1995, A Semiparametric Estimation Procedure of Dependence Parameters in Multivariate Families of Distributions, Biometrika 82, 543-552. Gorecki, J., Hofert, M. and Holena, M. 2014, On the Consistency of an Estimator for Hierarchical Archimedean Copulas, In Talaysova, J., Stoklasa, J., Talaysek, T. (Eds.) 32nd International Conference on Mathematical Methods in Economics, Olomouc: Palacky University, 239-244. Joe, H. 2005, Asymptotic Efficiency of the Two-Stage Estimation Method for Copula-Based Models, Journal of Multivariate Analysis 94(2), 401-419. Okhrin, O., Okhrin, Y. and Schmid, W. 2013, On the Structure and Estimation of Hierarchical Archimedean Copulas, Journal of Econometrics 173, 189-204. Okhrin, O. and Ristig, A. 2014, Hierarchical Archimedean Copulae: The Okhrin, O., Ristig, A., Sheen J. and Trueck, S. 2015, Conditional Systemic Risk with Penalized Copula, SFB 649 Discussion Paper 2015-038, Sonderforschungsbereich 649, Humboldt University, Germany. Examples
# define the copula model
tree = list(list("X1", "X5", 3), list("X2", "X3", "X4", 4), 2)
model = hac(type = 1, tree = tree)
# sample from copula model
x = rHAC(1000, model)
# in the following case the true model is binary approximated
est.obj = estimate.copula(x, type = 1, method = 1, epsilon = 0)
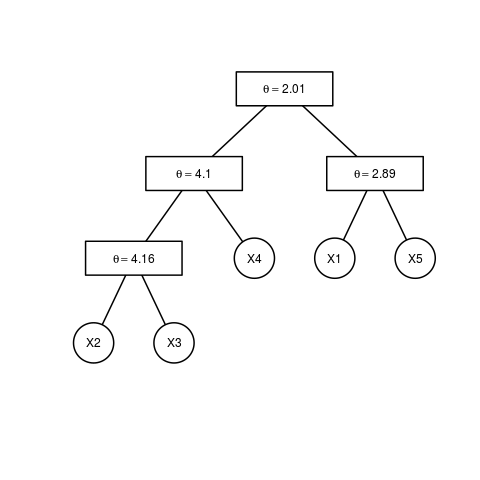
plot(est.obj)
# consider also the aggregation of the variables
est.obj = estimate.copula(x, type = 1, method = 1, epsilon = 0.2)
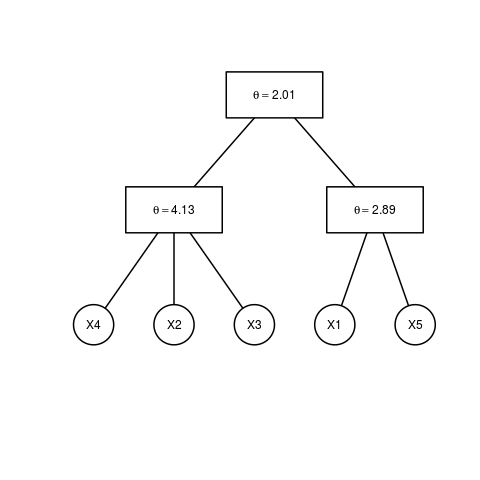
plot(est.obj)
# full ML estimation to yield more precise parameter
est.obj.full = estimate.copula(x, type = 1, method = 2, hac = est.obj)
# recursive ML estimation leads to almost identical results
est.obj.r = estimate.copula(x, type = 1, method = 3)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(HAC)
Loading required package: copula
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/HAC/estimate.copula.Rd_%03d_medium.png", width=480, height=480)
> ### Name: estimate.copula
> ### Title: Estimation of Hierarchical Archimedean Copulae
> ### Aliases: estimate.copula
>
> ### ** Examples
>
> # define the copula model
> tree = list(list("X1", "X5", 3), list("X2", "X3", "X4", 4), 2)
> model = hac(type = 1, tree = tree)
>
> # sample from copula model
> x = rHAC(1000, model)
[1] "X1 <-> 1"
[1] "X5 <-> 2"
[1] "X2 <-> 3"
[1] "X3 <-> 4"
[1] "X4 <-> 5"
Warning message:
In hac2nacopula(hac) : NAs introduced by coercion
>
> # in the following case the true model is binary approximated
> est.obj = estimate.copula(x, type = 1, method = 1, epsilon = 0)
> plot(est.obj)
>
> # consider also the aggregation of the variables
> est.obj = estimate.copula(x, type = 1, method = 1, epsilon = 0.2)
> plot(est.obj)
>
> # full ML estimation to yield more precise parameter
> est.obj.full = estimate.copula(x, type = 1, method = 2, hac = est.obj)
>
> # recursive ML estimation leads to almost identical results
> est.obj.r = estimate.copula(x, type = 1, method = 3)
>
>
>
>
>
> dev.off()
null device
1
>
|