|
Last data update: 2014.03.03
|
R: output and plot for ROR result
output and plot for ROR result
Description
function for output tables and figures related to ROR result
Usage
ODS.ror(case.sub, ctl.sub, lib.sub, lib.sub.names, records, dev.list, AIC.list, deleted.snps.ls, proteinf, locus = "DRB1*", ref.level = "101")
Arguments
case.sub |
case subjects, two columns for two haplotypes
|
ctl.sub |
control subjects, two columns for two haplotypes
|
lib.sub |
the alleles library contains allele sequences for those only appear in the case and control samples
|
lib.sub.names |
the corresponding names of the alleles (mapping of full name in the library and short name in samples)
|
records |
the record of the whole ROR process
|
dev.list |
deviances for all steps of ROR
|
AIC.list |
AICs for all steps of ROR
|
deleted.snps.ls |
the history of SNP deletions for all steps of ROR
|
proteinf |
amino acid matrix for the currsponding alleles
|
locus |
name of locus
|
ref.level |
name of reference allele
|
Author(s)
Xin Huang
References
Zhao, L.P. and Huang, X. Recursive organizer (ROR): an analytic framework for sequence-based association analysis. Human Genetics, 2013
Examples
library("HAP.ROR")
data(case.sub)
data(ctl.sub)
data(lib.sub)
data(lib.sub.names)
ror.res <- HAP.ror(case.sub, ctl.sub, lib.sub, lib.sub.names, alpha=0.01, ref.level="101");
# grouping result:
round(ror.res$dev.list, 2);
round(ror.res$AIC.list, 2);
ror.res$df.list;
ror.res$deleted.snps;
ror.res$grp.result;
ror.res$significant;
# model summary:
ror.res$model.summary;
# output tables and figures used for ror result
data(proteinf)
ODS.ror(case.sub=case.sub, ctl.sub=ctl.sub, lib.sub=lib.sub, lib.sub.names=lib.sub.names, records=ror.res$records, dev.list=ror.res$dev.list, AIC.list=ror.res$AIC.list, deleted.snps.ls=ror.res$deleted.snps.ls, proteinf=proteinf, locus="DRB1*", ref.level="101");
cat("ROR result tables/figures are store in folder:", getwd(),"\n")
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(HAP.ROR)
Loading required package: hash
hash-2.2.6 provided by Decision Patterns
Loading required package: ape
Attaching package: 'HAP.ROR'
The following object is masked from 'package:stats':
AIC
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/HAP.ROR/ODS.ror.Rd_%03d_medium.png", width=480, height=480)
> ### Name: ODS.ror
> ### Title: output and plot for ROR result
> ### Aliases: ODS.ror
> ### Keywords: ~kwd1 ~kwd2
>
> ### ** Examples
>
> library("HAP.ROR")
> data(case.sub)
> data(ctl.sub)
> data(lib.sub)
> data(lib.sub.names)
> ror.res <- HAP.ror(case.sub, ctl.sub, lib.sub, lib.sub.names, alpha=0.01, ref.level="101");
deleting: 49 50 51 61 68 70 76 78 80 81
deleting: 49 50 51 61 68 70 76 78 80 81 26 40 41 48 55 56 60 67 77 79
deleting: 49 50 51 61 68 70 76 78 80 81 26 40 41 48 55 56 60 67 77 79 12 23 25 29 52 57 59 62 63 65 66 69 71 72
deleting: 49 50 51 61 68 70 76 78 80 81 26 40 41 48 55 56 60 67 77 79 12 23 25 29 52 57 59 62 63 65 66 69 71 72 17 18 22 24 35 37 42 47 53 54 92
deleting: 49 50 51 61 68 70 76 78 80 81 26 40 41 48 55 56 60 67 77 79 12 23 25 29 52 57 59 62 63 65 66 69 71 72 17 18 22 24 35 37 42 47 53 54 92 1 2 4 7 13 14 15 16 39 83 85 86 89 90
deleting: 49 50 51 61 68 70 76 78 80 81 26 40 41 48 55 56 60 67 77 79 12 23 25 29 52 57 59 62 63 65 66 69 71 72 17 18 22 24 35 37 42 47 53 54 92 1 2 4 7 13 14 15 16 39 83 85 86 89 90 9 10 11 20 30 31 32 33 34 36 43 64 91
deleting: 49 50 51 61 68 70 76 78 80 81 26 40 41 48 55 56 60 67 77 79 12 23 25 29 52 57 59 62 63 65 66 69 71 72 17 18 22 24 35 37 42 47 53 54 92 1 2 4 7 13 14 15 16 39 83 85 86 89 90 9 10 11 20 30 31 32 33 34 36 43 64 91 8 58 74 75 87 88
deleting: 49 50 51 61 68 70 76 78 80 81 26 40 41 48 55 56 60 67 77 79 12 23 25 29 52 57 59 62 63 65 66 69 71 72 17 18 22 24 35 37 42 47 53 54 92 1 2 4 7 13 14 15 16 39 83 85 86 89 90 9 10 11 20 30 31 32 33 34 36 43 64 91 8 58 74 75 87 88 3 5 21 27 28 44 45 46 82 84
deleting: 49 50 51 61 68 70 76 78 80 81 26 40 41 48 55 56 60 67 77 79 12 23 25 29 52 57 59 62 63 65 66 69 71 72 17 18 22 24 35 37 42 47 53 54 92 1 2 4 7 13 14 15 16 39 83 85 86 89 90 9 10 11 20 30 31 32 33 34 36 43 64 91 8 58 74 75 87 88 3 5 21 27 28 44 45 46 82 84 6 19 38 73
There were 12 warnings (use warnings() to see them)
>
> # grouping result:
> round(ror.res$dev.list, 2);
[1] 0.00 0.02 4.29 3.51 3.75 0.21 4.90 3.32 -91.25
> round(ror.res$AIC.list, 2);
[1] 0.95 0.88 0.04 0.06 0.05 0.65 0.03 0.07 0.00
> ror.res$df.list;
[1] 1 1 1 1 1 1 1 1 -75
> ror.res$deleted.snps;
[1] 1 2 3 4 5 7 8 9 10 11 12 13 14 15 16 17 18 20 21 22 23 24 25 26 27
[26] 28 29 30 31 32 33 34 35 36 37 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53
[51] 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 74 75 76 77 78 79
[76] 80 81 82 83 84 85 86 87 88 89 90 91 92
> ror.res$grp.result;
grp
[1,] "DRB1*01:01" "1"
[2,] "DRB1*03:01" "1"
[3,] "DRB1*04:01" "1"
[4,] "DRB1*04:04" "1"
[5,] "DRB1*07:01" "1"
[6,] "DRB1*08:01" "1"
[7,] "DRB1*09:01" "1"
[8,] "DRB1*13:01" "1"
[9,] "DRB1*13:02" "1"
> ror.res$significant;
[1] 1
> # model summary:
> ror.res$model.summary;
Call:
glm(formula = grp.sample[, 1] ~ allele.factor, family = binomial)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.435 -1.435 0.940 0.940 0.940
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.5878 0.2494 2.356 0.0185 *
allele.factor -18.2099 1819.5131 -0.010 0.9920
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 104.539 on 76 degrees of freedom
Residual deviance: 91.246 on 75 degrees of freedom
AIC: 95.246
Number of Fisher Scoring iterations: 17
> # output tables and figures used for ror result
> data(proteinf)
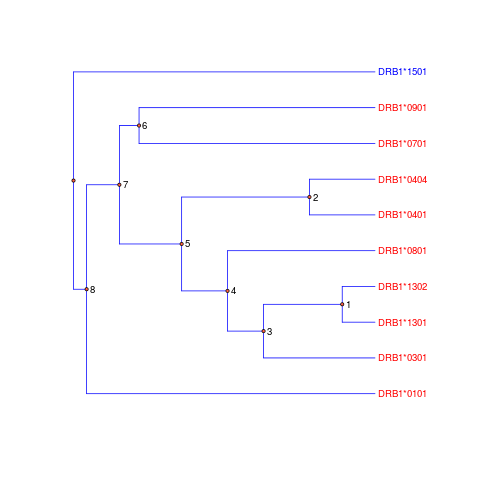
> ODS.ror(case.sub=case.sub, ctl.sub=ctl.sub, lib.sub=lib.sub, lib.sub.names=lib.sub.names, records=ror.res$records, dev.list=ror.res$dev.list, AIC.list=ror.res$AIC.list, deleted.snps.ls=ror.res$deleted.snps.ls, proteinf=proteinf, locus="DRB1*", ref.level="101");
Warning messages:
1: glm.fit: fitted probabilities numerically 0 or 1 occurred
2: glm.fit: fitted probabilities numerically 0 or 1 occurred
3: glm.fit: fitted probabilities numerically 0 or 1 occurred
4: glm.fit: fitted probabilities numerically 0 or 1 occurred
5: glm.fit: fitted probabilities numerically 0 or 1 occurred
6: glm.fit: fitted probabilities numerically 0 or 1 occurred
7: glm.fit: fitted probabilities numerically 0 or 1 occurred
8: glm.fit: fitted probabilities numerically 0 or 1 occurred
> cat("ROR result tables/figures are store in folder:", getwd(),"\n")
ROR result tables/figures are store in folder: /home/ddbj/DataUpdator-rgm3/target
>
>
>
>
>
> dev.off()
null device
1
>
|
|
 providing
providing  .
.