Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Additive Regression with Optimal Transformations on Both Sides using Canonical VariatesDescriptionExpands continuous variables into restricted cubic spline bases and
categorical variables into dummy variables and fits a multivariate
equation using canonical variates. This finds optimum transformations
that maximize R^2. Optionally, the bootstrap is used to estimate
the covariance matrix of both left- and right-hand-side transformation
parameters, and to estimate the bias in the R^2 due to overfitting
and compute the bootstrap optimism-corrected R^2.
Cross-validation can also be used to get an unbiased estimate of
R^2 but this is not as precise as the bootstrap estimate. The
bootstrap and cross-validation may also used to get estimates of mean
and median absolute error in predicted values on the original Note that uncertainty about the proper transformation of Usage
areg(x, y, xtype = NULL, ytype = NULL, nk = 4,
B = 0, na.rm = TRUE, tolerance = NULL, crossval = NULL)
## S3 method for class 'areg'
print(x, digits=4, ...)
## S3 method for class 'areg'
plot(x, whichx = 1:ncol(x$x), ...)
## S3 method for class 'areg'
predict(object, x, type=c('lp','fitted','x'),
what=c('all','sample'), ...)
Arguments
Details
If one side of the equation has a categorical variable with more than two categories and the other side has a continuous variable not assumed to act linearly, larger sample sizes are needed to reliably estimate transformations, as it is difficult to optimally score categorical variables to maximize R^2 against a simultaneously optimally transformed continuous variable. Valuea list of class Author(s)Frank Harrell
ReferencesBreiman and Friedman, Journal of the American Statistical Association (September, 1985). See Also
Examples
set.seed(1)
ns <- c(30,300,3000)
for(n in ns) {
y <- sample(1:5, n, TRUE)
x <- abs(y-3) + runif(n)
par(mfrow=c(3,4))
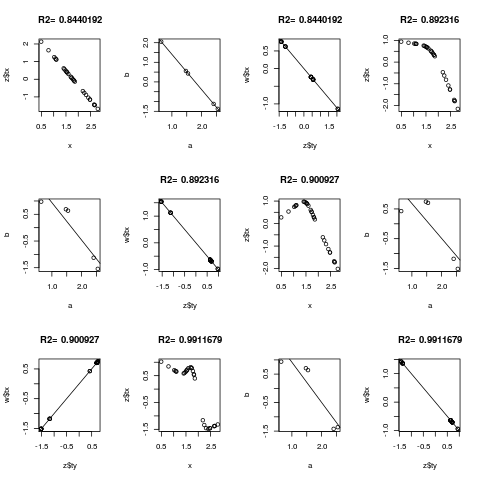
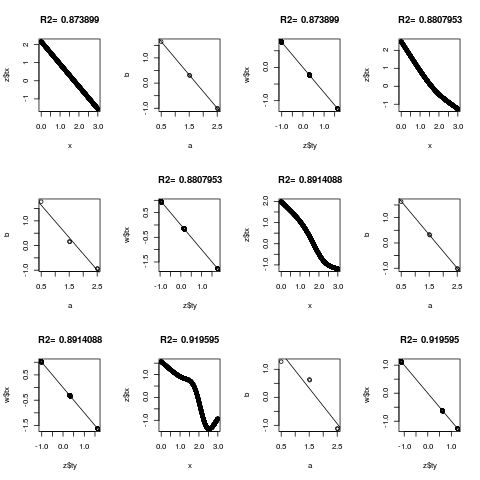
for(k in c(0,3:5)) {
z <- areg(x, y, ytype='c', nk=k)
plot(x, z$tx)
title(paste('R2=',format(z$rsquared)))
tapply(z$ty, y, range)
a <- tapply(x,y,mean)
b <- tapply(z$ty,y,mean)
plot(a,b)
abline(lsfit(a,b))
# Should get same result to within linear transformation if reverse x and y
w <- areg(y, x, xtype='c', nk=k)
plot(z$ty, w$tx)
title(paste('R2=',format(w$rsquared)))
abline(lsfit(z$ty, w$tx))
}
}
par(mfrow=c(2,2))
# Example where one category in y differs from others but only in variance of x
n <- 50
y <- sample(1:5,n,TRUE)
x <- rnorm(n)
x[y==1] <- rnorm(sum(y==1), 0, 5)
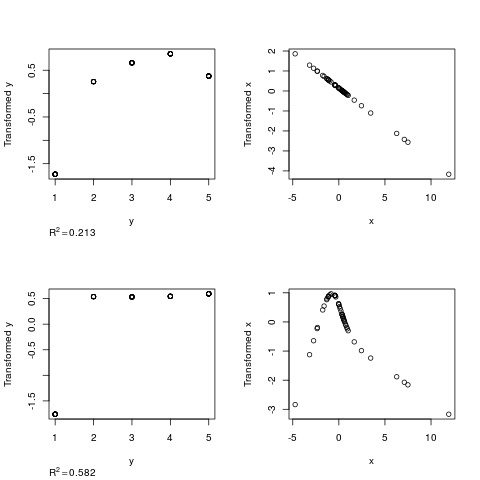
z <- areg(x,y,xtype='l',ytype='c')
z
plot(z)
z <- areg(x,y,ytype='c')
z
plot(z)
## Not run:
# Examine overfitting when true transformations are linear
par(mfrow=c(4,3))
for(n in c(200,2000)) {
x <- rnorm(n); y <- rnorm(n) + x
for(nk in c(0,3,5)) {
z <- areg(x, y, nk=nk, crossval=10, B=100)
print(z)
plot(z)
title(paste('n=',n))
}
}
par(mfrow=c(1,1))
# Underfitting when true transformation is quadratic but overfitting
# when y is allowed to be transformed
set.seed(49)
n <- 200
x <- rnorm(n); y <- rnorm(n) + .5*x^2
#areg(x, y, nk=0, crossval=10, B=100)
#areg(x, y, nk=4, ytype='l', crossval=10, B=100)
z <- areg(x, y, nk=4) #, crossval=10, B=100)
z
# Plot x vs. predicted value on original scale. Since y-transform is
# not monotonic, there are multiple y-inverses
xx <- seq(-3.5,3.5,length=1000)
yhat <- predict(z, xx, type='fitted')
plot(x, y, xlim=c(-3.5,3.5))
for(j in 1:ncol(yhat)) lines(xx, yhat[,j], col=j)
# Plot a random sample of possible y inverses
yhats <- predict(z, xx, type='fitted', what='sample')
points(xx, yhats, pch=2)
## End(Not run)
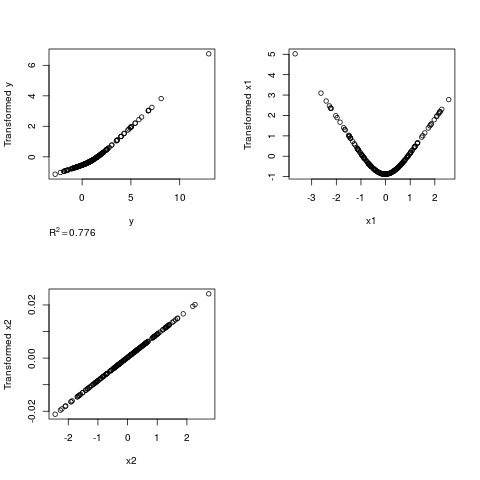
# True transformation of x1 is quadratic, y is linear
n <- 200
x1 <- rnorm(n); x2 <- rnorm(n); y <- rnorm(n) + x1^2
z <- areg(cbind(x1,x2),y,xtype=c('s','l'),nk=3)
par(mfrow=c(2,2))
plot(z)
# y transformation is inverse quadratic but areg gets the same answer by
# making x1 quadratic
n <- 5000
x1 <- rnorm(n); x2 <- rnorm(n); y <- (x1 + rnorm(n))^2
z <- areg(cbind(x1,x2),y,nk=5)
par(mfrow=c(2,2))
plot(z)
# Overfit 20 predictors when no true relationships exist
n <- 1000
x <- matrix(runif(n*20),n,20)
y <- rnorm(n)
z <- areg(x, y, nk=5) # add crossval=4 to expose the problem
# Test predict function
n <- 50
x <- rnorm(n)
y <- rnorm(n) + x
g <- sample(1:3, n, TRUE)
z <- areg(cbind(x,g),y,xtype=c('s','c'))
range(predict(z, cbind(x,g)) - z$linear.predictors)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(Hmisc)
Loading required package: lattice
Loading required package: survival
Loading required package: Formula
Loading required package: ggplot2
Attaching package: 'Hmisc'
The following objects are masked from 'package:base':
format.pval, round.POSIXt, trunc.POSIXt, units
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/Hmisc/areg.Rd_%03d_medium.png", width=480, height=480)
> ### Name: areg
> ### Title: Additive Regression with Optimal Transformations on Both Sides
> ### using Canonical Variates
> ### Aliases: areg print.areg predict.areg plot.areg
> ### Keywords: smooth regression multivariate models
>
> ### ** Examples
>
> set.seed(1)
>
> ns <- c(30,300,3000)
> for(n in ns) {
+ y <- sample(1:5, n, TRUE)
+ x <- abs(y-3) + runif(n)
+ par(mfrow=c(3,4))
+ for(k in c(0,3:5)) {
+ z <- areg(x, y, ytype='c', nk=k)
+ plot(x, z$tx)
+ title(paste('R2=',format(z$rsquared)))
+ tapply(z$ty, y, range)
+ a <- tapply(x,y,mean)
+ b <- tapply(z$ty,y,mean)
+ plot(a,b)
+ abline(lsfit(a,b))
+ # Should get same result to within linear transformation if reverse x and y
+ w <- areg(y, x, xtype='c', nk=k)
+ plot(z$ty, w$tx)
+ title(paste('R2=',format(w$rsquared)))
+ abline(lsfit(z$ty, w$tx))
+ }
+ }
>
> par(mfrow=c(2,2))
> # Example where one category in y differs from others but only in variance of x
> n <- 50
> y <- sample(1:5,n,TRUE)
> x <- rnorm(n)
> x[y==1] <- rnorm(sum(y==1), 0, 5)
> z <- areg(x,y,xtype='l',ytype='c')
> z
N: 50 0 observations with NAs deleted.
R^2: 0.213 nk: 4 Mean and Median |error|: 2.06, 2
type d.f.
x l 1
y type: c d.f.: 4
> plot(z)
> z <- areg(x,y,ytype='c')
> z
N: 50 0 observations with NAs deleted.
R^2: 0.582 nk: 4 Mean and Median |error|: 2.06, 2
type d.f.
x s 3
y type: c d.f.: 4
> plot(z)
>
> ## Not run:
> ##D
> ##D # Examine overfitting when true transformations are linear
> ##D par(mfrow=c(4,3))
> ##D for(n in c(200,2000)) {
> ##D x <- rnorm(n); y <- rnorm(n) + x
> ##D for(nk in c(0,3,5)) {
> ##D z <- areg(x, y, nk=nk, crossval=10, B=100)
> ##D print(z)
> ##D plot(z)
> ##D title(paste('n=',n))
> ##D }
> ##D }
> ##D par(mfrow=c(1,1))
> ##D
> ##D # Underfitting when true transformation is quadratic but overfitting
> ##D # when y is allowed to be transformed
> ##D set.seed(49)
> ##D n <- 200
> ##D x <- rnorm(n); y <- rnorm(n) + .5*x^2
> ##D #areg(x, y, nk=0, crossval=10, B=100)
> ##D #areg(x, y, nk=4, ytype='l', crossval=10, B=100)
> ##D z <- areg(x, y, nk=4) #, crossval=10, B=100)
> ##D z
> ##D # Plot x vs. predicted value on original scale. Since y-transform is
> ##D # not monotonic, there are multiple y-inverses
> ##D xx <- seq(-3.5,3.5,length=1000)
> ##D yhat <- predict(z, xx, type='fitted')
> ##D plot(x, y, xlim=c(-3.5,3.5))
> ##D for(j in 1:ncol(yhat)) lines(xx, yhat[,j], col=j)
> ##D # Plot a random sample of possible y inverses
> ##D yhats <- predict(z, xx, type='fitted', what='sample')
> ##D points(xx, yhats, pch=2)
> ## End(Not run)
>
> # True transformation of x1 is quadratic, y is linear
> n <- 200
> x1 <- rnorm(n); x2 <- rnorm(n); y <- rnorm(n) + x1^2
> z <- areg(cbind(x1,x2),y,xtype=c('s','l'),nk=3)
> par(mfrow=c(2,2))
> plot(z)
>
> # y transformation is inverse quadratic but areg gets the same answer by
> # making x1 quadratic
> n <- 5000
> x1 <- rnorm(n); x2 <- rnorm(n); y <- (x1 + rnorm(n))^2
> z <- areg(cbind(x1,x2),y,nk=5)
> par(mfrow=c(2,2))
> plot(z)
>
> # Overfit 20 predictors when no true relationships exist
> n <- 1000
> x <- matrix(runif(n*20),n,20)
> y <- rnorm(n)
> z <- areg(x, y, nk=5) # add crossval=4 to expose the problem
>
> # Test predict function
> n <- 50
> x <- rnorm(n)
> y <- rnorm(n) + x
> g <- sample(1:3, n, TRUE)
> z <- areg(cbind(x,g),y,xtype=c('s','c'))
> range(predict(z, cbind(x,g)) - z$linear.predictors)
[1] 0 0
>
>
>
>
>
> dev.off()
null device
1
>
|