Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Rank Correlation for Censored DataDescriptionComputes the c index and the corresponding generalization of Somers' Dxy rank correlation for a censored response variable. Also works for uncensored and binary responses, although its use of all possible pairings makes it slow for this purpose. Dxy and c are related by var{Dxy} = 2*(var{c} - 0.5).
Usage
rcorr.cens(x, S, outx=FALSE)
## S3 method for class 'formula'
rcorrcens(formula, data=NULL, subset=NULL,
na.action=na.retain, exclude.imputed=TRUE, outx=FALSE,
...)
Arguments
Value
Author(s)Frank Harrell
ReferencesNewson R: Confidence intervals for rank statistics: Somers' D and extensions. Stata Journal 6:309-334; 2006. See Also
Examples
set.seed(1)
x <- round(rnorm(200))
y <- rnorm(200)
rcorr.cens(x, y, outx=TRUE) # can correlate non-censored variables
library(survival)
age <- rnorm(400, 50, 10)
bp <- rnorm(400,120, 15)
bp[1] <- NA
d.time <- rexp(400)
cens <- runif(400,.5,2)
death <- d.time <= cens
d.time <- pmin(d.time, cens)
rcorr.cens(age, Surv(d.time, death))
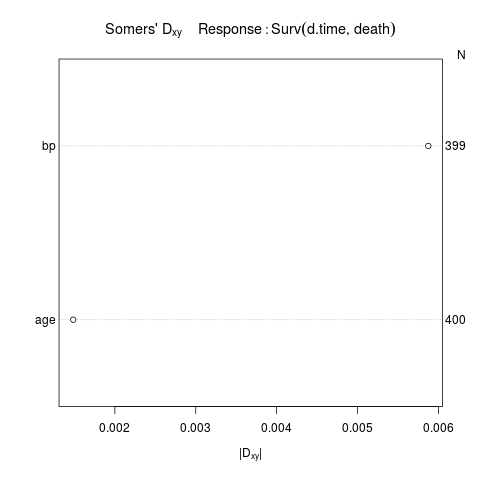
r <- rcorrcens(Surv(d.time, death) ~ age + bp)
r
plot(r)
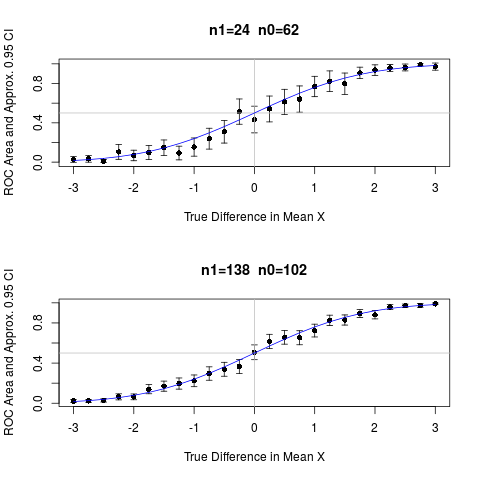
# Show typical 0.95 confidence limits for ROC areas for a sample size
# with 24 events and 62 non-events, for varying population ROC areas
# Repeat for 138 events and 102 non-events
set.seed(8)
par(mfrow=c(2,1))
for(i in 1:2) {
n1 <- c(24,138)[i]
n0 <- c(62,102)[i]
y <- c(rep(0,n0), rep(1,n1))
deltas <- seq(-3, 3, by=.25)
C <- se <- deltas
j <- 0
for(d in deltas) {
j <- j + 1
x <- c(rnorm(n0, 0), rnorm(n1, d))
w <- rcorr.cens(x, y)
C[j] <- w['C Index']
se[j] <- w['S.D.']/2
}
low <- C-1.96*se; hi <- C+1.96*se
print(cbind(C, low, hi))
errbar(deltas, C, C+1.96*se, C-1.96*se,
xlab='True Difference in Mean X',
ylab='ROC Area and Approx. 0.95 CI')
title(paste('n1=',n1,' n0=',n0,sep=''))
abline(h=.5, v=0, col='gray')
true <- 1 - pnorm(0, deltas, sqrt(2))
lines(deltas, true, col='blue')
}
par(mfrow=c(1,1))
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(Hmisc)
Loading required package: lattice
Loading required package: survival
Loading required package: Formula
Loading required package: ggplot2
Attaching package: 'Hmisc'
The following objects are masked from 'package:base':
format.pval, round.POSIXt, trunc.POSIXt, units
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/Hmisc/rcorr.cens.Rd_%03d_medium.png", width=480, height=480)
> ### Name: rcorr.cens
> ### Title: Rank Correlation for Censored Data
> ### Aliases: rcorr.cens rcorrcens rcorrcens.formula
> ### Keywords: survival nonparametric
>
> ### ** Examples
>
> set.seed(1)
> x <- round(rnorm(200))
> y <- rnorm(200)
> rcorr.cens(x, y, outx=TRUE) # can correlate non-censored variables
C Index Dxy S.D. n missing
4.762913e-01 -4.741744e-02 6.488578e-02 2.000000e+02 0.000000e+00
uncensored Relevant Pairs Concordant Uncertain
2.000000e+02 2.834400e+04 1.350000e+04 0.000000e+00
> library(survival)
> age <- rnorm(400, 50, 10)
> bp <- rnorm(400,120, 15)
> bp[1] <- NA
> d.time <- rexp(400)
> cens <- runif(400,.5,2)
> death <- d.time <= cens
> d.time <- pmin(d.time, cens)
> rcorr.cens(age, Surv(d.time, death))
C Index Dxy S.D. n missing
4.992573e-01 -1.485304e-03 3.673749e-02 4.000000e+02 0.000000e+00
uncensored Relevant Pairs Concordant Uncertain
2.790000e+02 1.333060e+05 6.655400e+04 2.629400e+04
> r <- rcorrcens(Surv(d.time, death) ~ age + bp)
> r
Somers' Rank Correlation for Censored Data Response variable:Surv(d.time, death)
C Dxy aDxy SD Z P n
age 0.499 -0.001 0.001 0.037 0.04 0.9678 400
bp 0.497 -0.006 0.006 0.038 0.15 0.8773 399
> plot(r)
>
> # Show typical 0.95 confidence limits for ROC areas for a sample size
> # with 24 events and 62 non-events, for varying population ROC areas
> # Repeat for 138 events and 102 non-events
> set.seed(8)
> par(mfrow=c(2,1))
> for(i in 1:2) {
+ n1 <- c(24,138)[i]
+ n0 <- c(62,102)[i]
+ y <- c(rep(0,n0), rep(1,n1))
+ deltas <- seq(-3, 3, by=.25)
+ C <- se <- deltas
+ j <- 0
+ for(d in deltas) {
+ j <- j + 1
+ x <- c(rnorm(n0, 0), rnorm(n1, d))
+ w <- rcorr.cens(x, y)
+ C[j] <- w['C Index']
+ se[j] <- w['S.D.']/2
+ }
+ low <- C-1.96*se; hi <- C+1.96*se
+ print(cbind(C, low, hi))
+ errbar(deltas, C, C+1.96*se, C-1.96*se,
+ xlab='True Difference in Mean X',
+ ylab='ROC Area and Approx. 0.95 CI')
+ title(paste('n1=',n1,' n0=',n0,sep=''))
+ abline(h=.5, v=0, col='gray')
+ true <- 1 - pnorm(0, deltas, sqrt(2))
+ lines(deltas, true, col='blue')
+ }
C low hi
[1,] 0.02755376 -0.0015097885 0.05661732
[2,] 0.03360215 -0.0003784209 0.06758272
[3,] 0.01075269 -0.0044497156 0.02595509
[4,] 0.10416667 0.0284823228 0.17985101
[5,] 0.06787634 0.0142995376 0.12145315
[6,] 0.09811828 0.0262850181 0.16995154
[7,] 0.14717742 0.0687615968 0.22559324
[8,] 0.09274194 0.0231499208 0.16233395
[9,] 0.15322581 0.0606689017 0.24578271
[10,] 0.23857527 0.1324745070 0.34467603
[11,] 0.30846774 0.1940638328 0.42287165
[12,] 0.51411290 0.3856972238 0.64252858
[13,] 0.43346774 0.2978050069 0.56913048
[14,] 0.54099462 0.4094129537 0.67257629
[15,] 0.61357527 0.4861716011 0.74097894
[16,] 0.64180108 0.5083995705 0.77520258
[17,] 0.76948925 0.6660036397 0.87297485
[18,] 0.82325269 0.7183521941 0.92815318
[19,] 0.79771505 0.6891303373 0.90629977
[20,] 0.90793011 0.8496264055 0.96623381
[21,] 0.93615591 0.8826444375 0.98966739
[22,] 0.95766129 0.9218631555 0.99345943
[23,] 0.96303763 0.9280097068 0.99806556
[24,] 0.99059140 0.9780487323 1.00313406
[25,] 0.97177419 0.9353647894 1.00818360
C low hi
[1,] 0.02195226 0.006435429 0.03746909
[2,] 0.02394146 0.009333689 0.03854923
[3,] 0.02791986 0.011615456 0.04422427
[4,] 0.06351236 0.033414116 0.09361061
[5,] 0.06344132 0.034476786 0.09240585
[6,] 0.13981245 0.094242001 0.18538289
[7,] 0.17014777 0.119801626 0.22049391
[8,] 0.19607843 0.140330050 0.25182681
[9,] 0.22300369 0.164653945 0.28135344
[10,] 0.29560955 0.228818159 0.36240094
[11,] 0.33823529 0.268816721 0.40765387
[12,] 0.36608411 0.295661218 0.43650701
[13,] 0.50745951 0.433680623 0.58123839
[14,] 0.61530264 0.544122164 0.68648312
[15,] 0.65686275 0.588520795 0.72520469
[16,] 0.65231600 0.582394897 0.72223710
[17,] 0.72371412 0.659942480 0.78748577
[18,] 0.82551861 0.773470052 0.87756717
[19,] 0.82914180 0.778668043 0.87961556
[20,] 0.89386189 0.854440532 0.93328325
[21,] 0.88128730 0.840157819 0.92241678
[22,] 0.95865303 0.935219169 0.98208688
[23,] 0.97158284 0.954699265 0.98846641
[24,] 0.97428247 0.956007156 0.99255778
[25,] 0.98991191 0.981473900 0.99834991
> par(mfrow=c(1,1))
>
>
>
>
>
> dev.off()
null device
1
>
|