Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Create survival curves based on the IDP modelDescriptionThis function creates survival curves from right censored data using the prior near-ignorance Dirichlet Process (IDP). Usage
isurvfit(formula, data, s=0.5, weights, subset, display=TRUE,
conf.type=c('exact', 'approx', 'none'), nsamples=10000,
conf.int= .95)
Arguments
DetailsThe estimates are obtained using the IDP estimator by Mangili and others (2014) based on the prior near-ignorance Dirichlet Process model by Benavoli and others (2014). Valuean object of class See ReferencesBenavoli, A., Mangili, F., Zaffalon, M. and Ruggeri, F. (2014). Imprecise Dirichlet process with application to the hypothesis test on the probability that X < Y. ArXiv e-prints, http://adsabs.harvard.edu/abs/2014arXiv1402.2755B. Mangili, F., Benavoli, A., Zaffalon, M. and de Campos, C. (2014). Imprecise Dirichlet Process for the estimate and comparison of survival functions with censored data. See Also
Examples
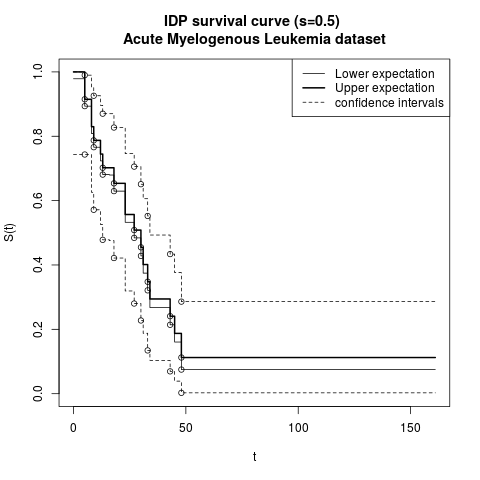
data(aml)
fit <- isurvfit(Surv(time, cens) ~ 1, data=aml, display=TRUE, nsamples=1000)
legend('topright', c("Lower expectation",
"Upper expectation","confidence intervals"), lty=c(1,1,2),lwd=c(1,2,1))
title("IDP survival curve (s=0.5) \nAcute Myelogenous Leukemia dataset")
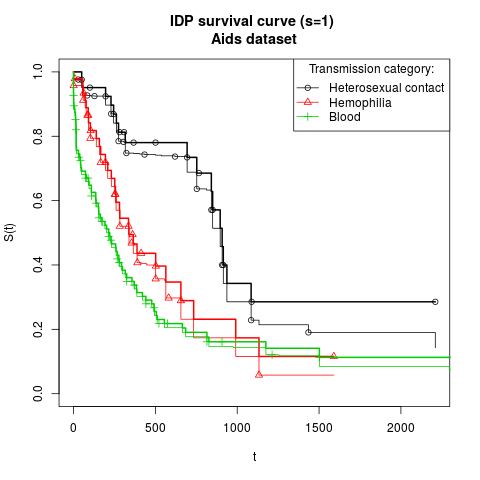
data(Aids2)
dataset <- Aids2
dataset["time"]<-dataset[4]-dataset[3]
dataset[5]<-as.numeric(unlist(dataset[5]))
fit <- isurvfit(Surv(time, status) ~ T.categ, dataset,s=1,
subset=(!is.na(match(T.categ, c('blood','haem','het')))),
nsamples=1000,conf.type='none')
legend('topright',c("Heterosexual contact","Hemophilia","Blood"),
title="Transmission category:",lty=c(1,1,1),col=c(1,2,3),pch=c(1,2,3))
title("IDP survival curve (s=1) \nAids dataset")
print(fit)
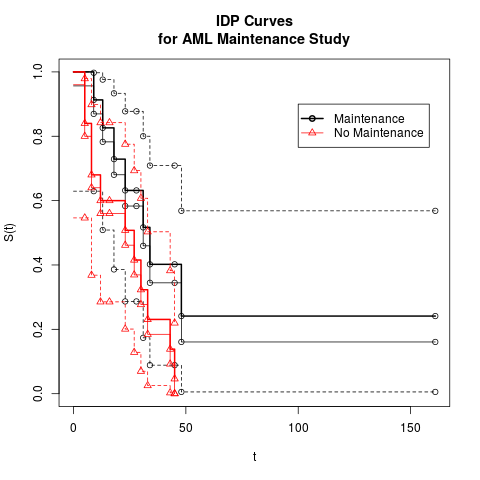
leukemia.surv <- isurvfit(Surv(time, cens) ~ group, data = aml, display=FALSE)
plot(leukemia.surv)
legend(100, .9, c("Maintenance", "No Maintenance"), lty=c(1,1),lwd=c(2,1),
col=c('black','red'),pch=c(1,2))
title("IDP Curves\nfor AML Maintenance Study")
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(IDPSurvival)
Loading required package: Rsolnp
Loading required package: gtools
Loading required package: survival
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/IDPSurvival/isurvfit.Rd_%03d_medium.png", width=480, height=480)
> ### Name: isurvfit
> ### Title: Create survival curves based on the IDP model
> ### Aliases: isurvfit print.isurvfit
> ### Keywords: survival IDP
>
> ### ** Examples
>
> data(aml)
> fit <- isurvfit(Surv(time, cens) ~ 1, data=aml, display=TRUE, nsamples=1000)
> legend('topright', c("Lower expectation",
+ "Upper expectation","confidence intervals"), lty=c(1,1,2),lwd=c(1,2,1))
> title("IDP survival curve (s=0.5) \nAcute Myelogenous Leukemia dataset")
>
> data(Aids2)
> dataset <- Aids2
> dataset["time"]<-dataset[4]-dataset[3]
> dataset[5]<-as.numeric(unlist(dataset[5]))
> fit <- isurvfit(Surv(time, status) ~ T.categ, dataset,s=1,
+ subset=(!is.na(match(T.categ, c('blood','haem','het')))),
+ nsamples=1000,conf.type='none')
> legend('topright',c("Heterosexual contact","Hemophilia","Blood"),
+ title="Transmission category:",lty=c(1,1,1),col=c(1,2,3),pch=c(1,2,3))
> title("IDP survival curve (s=1) \nAids dataset")
> print(fit)
n.records n.events n.censored
T.categ=het 41 17 24
T.categ=haem 46 29 17
T.categ=blood 78 76 18
>
> leukemia.surv <- isurvfit(Surv(time, cens) ~ group, data = aml, display=FALSE)
> plot(leukemia.surv)
> legend(100, .9, c("Maintenance", "No Maintenance"), lty=c(1,1),lwd=c(2,1),
+ col=c('black','red'),pch=c(1,2))
> title("IDP Curves\nfor AML Maintenance Study")
>
>
>
>
>
> dev.off()
null device
1
>
|