Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Quick computation of kriging covariancesDescriptionComputes kriging covariances between one new point and many integration points, using precomputed data. UsagecomputeQuickKrigcov(model,integration.points,X.new, precalc.data, F.newdata , c.newdata) Arguments
DetailsThis function requires to use another function in order to generate the proper arguments.
The argument ValueA vector containing kriging covariances Author(s)Clement Chevalier (IMSV, Switzerland, and IRSN, France) ReferencesChevalier C., Bect J., Ginsbourger D., Vazquez E., Picheny V., Richet Y. (2011), Fast parallel kriging-based stepwise uncertainty reduction with application to the identification of an excursion set ,http://hal.archives-ouvertes.fr/hal-00641108/ Chevalier C., Ginsbourger D. (2012), Corrected Kriging update formulae for batch-sequential data assimilation ,http://arxiv.org/pdf/1203.6452.pdf See Also
Examples
#computeQuickKrigcov
set.seed(8)
N <- 9 #number of observations
testfun <- branin
#a 9 points initial design
design <- data.frame( matrix(runif(2*N),ncol=2) )
response <- testfun(design)
#km object with matern3_2 covariance
#params estimated by ML from the observations
model <- km(formula=~., design = design,
response = response,covtype="matern3_2")
#the points where we want to compute prediction
#if a point new.x is added to the doe
n.grid <- 20 #you can run it with 100
x.grid <- y.grid <- seq(0,1,length=n.grid)
newdata <- expand.grid(x.grid,y.grid)
#precalculation
precalc.data <- precomputeUpdateData(model=model,
integration.points=newdata)
#now we can compute very quickly kriging covariances
#between these data and any other points
other.x <- matrix(c(0.6,0.6),ncol=2)
pred <- predict_nobias_km(object=model,
newdata=other.x,type="UK",se.compute=TRUE)
kn <- computeQuickKrigcov(model=model,
integration.points=newdata,X.new=other.x,
precalc.data=precalc.data,
F.newdata=pred$F.newdata,
c.newdata=pred$c)
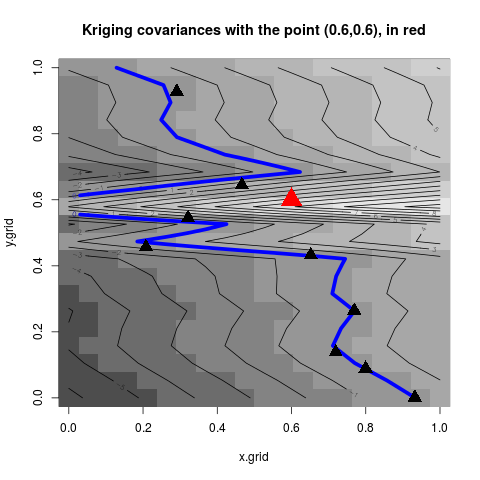
z.grid <- matrix(kn, n.grid, n.grid)
#plots: contour of the criterion, doe points and new point
image(x=x.grid,y=y.grid,z=z.grid,col=grey.colors(10))
contour(x=x.grid,y=y.grid,z=z.grid,15,add=TRUE)
contour(x=x.grid,y=y.grid,z=z.grid,levels=0,add=TRUE,col="blue",lwd=5)
points(design, col="black", pch=17, lwd=4,cex=2)
points(other.x, col="red", pch=17, lwd=4,cex=3)
title("Kriging covariances with the point (0.6,0.6), in red")
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(KrigInv)
Loading required package: DiceKriging
Loading required package: pbivnorm
Loading required package: rgenoud
## rgenoud (Version 5.7-12.4, Build Date: 2015-07-19)
## See http://sekhon.berkeley.edu/rgenoud for additional documentation.
## Please cite software as:
## Walter Mebane, Jr. and Jasjeet S. Sekhon. 2011.
## ``Genetic Optimization Using Derivatives: The rgenoud package for R.''
## Journal of Statistical Software, 42(11): 1-26.
##
Loading required package: randtoolbox
Loading required package: rngWELL
This is randtoolbox. For overview, type 'help("randtoolbox")'.
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/KrigInv/computeQuickKrigcov.Rd_%03d_medium.png", width=480, height=480)
> ### Name: computeQuickKrigcov
> ### Title: Quick computation of kriging covariances
> ### Aliases: computeQuickKrigcov
>
> ### ** Examples
>
> #computeQuickKrigcov
>
> set.seed(8)
> N <- 9 #number of observations
> testfun <- branin
>
> #a 9 points initial design
> design <- data.frame( matrix(runif(2*N),ncol=2) )
> response <- testfun(design)
>
> #km object with matern3_2 covariance
> #params estimated by ML from the observations
> model <- km(formula=~., design = design,
+ response = response,covtype="matern3_2")
optimisation start
------------------
* estimation method : MLE
* optimisation method : BFGS
* analytical gradient : used
* trend model : ~X1 + X2
* covariance model :
- type : matern3_2
- nugget : NO
- parameters lower bounds : 1e-10 1e-10
- parameters upper bounds : 1.448893 1.853021
- best initial criterion value(s) : -25.38168
N = 2, M = 5 machine precision = 2.22045e-16
At X0, 0 variables are exactly at the bounds
At iterate 0 f= 25.382 |proj g|= 0.19431
At iterate 1 f = 25.027 |proj g|= 0.13259
At iterate 2 f = 25.014 |proj g|= 1.6725
At iterate 3 f = 25.002 |proj g|= 0.15969
At iterate 4 f = 25.001 |proj g|= 0.17792
At iterate 5 f = 24.999 |proj g|= 0.31318
At iterate 6 f = 24.998 |proj g|= 0.14968
At iterate 7 f = 24.998 |proj g|= 0.03446
At iterate 8 f = 24.998 |proj g|= 0.03458
At iterate 9 f = 24.998 |proj g|= 0.0084816
At iterate 10 f = 24.998 |proj g|= 0.038393
At iterate 11 f = 24.997 |proj g|= 1.3196
At iterate 12 f = 24.997 |proj g|= 1.3327
At iterate 13 f = 24.994 |proj g|= 1.8077
At iterate 14 f = 24.991 |proj g|= 1.8106
At iterate 15 f = 24.975 |proj g|= 1.8136
At iterate 16 f = 24.937 |proj g|= 1.8202
At iterate 17 f = 24.816 |proj g|= 1.8136
At iterate 18 f = 24.652 |proj g|= 0.81261
At iterate 19 f = 24.652 |proj g|= 0.25743
At iterate 20 f = 24.651 |proj g|= 0.0033442
At iterate 21 f = 24.651 |proj g|= 1.4045e-05
iterations 21
function evaluations 30
segments explored during Cauchy searches 22
BFGS updates skipped 0
active bounds at final generalized Cauchy point 1
norm of the final projected gradient 1.40447e-05
final function value 24.6515
F = 24.6515
final value 24.651471
converged
>
> #the points where we want to compute prediction
> #if a point new.x is added to the doe
> n.grid <- 20 #you can run it with 100
> x.grid <- y.grid <- seq(0,1,length=n.grid)
> newdata <- expand.grid(x.grid,y.grid)
>
> #precalculation
> precalc.data <- precomputeUpdateData(model=model,
+ integration.points=newdata)
>
> #now we can compute very quickly kriging covariances
> #between these data and any other points
> other.x <- matrix(c(0.6,0.6),ncol=2)
> pred <- predict_nobias_km(object=model,
+ newdata=other.x,type="UK",se.compute=TRUE)
>
> kn <- computeQuickKrigcov(model=model,
+ integration.points=newdata,X.new=other.x,
+ precalc.data=precalc.data,
+ F.newdata=pred$F.newdata,
+ c.newdata=pred$c)
>
> z.grid <- matrix(kn, n.grid, n.grid)
>
> #plots: contour of the criterion, doe points and new point
> image(x=x.grid,y=y.grid,z=z.grid,col=grey.colors(10))
> contour(x=x.grid,y=y.grid,z=z.grid,15,add=TRUE)
> contour(x=x.grid,y=y.grid,z=z.grid,levels=0,add=TRUE,col="blue",lwd=5)
> points(design, col="black", pch=17, lwd=4,cex=2)
> points(other.x, col="red", pch=17, lwd=4,cex=3)
> title("Kriging covariances with the point (0.6,0.6), in red")
>
>
>
>
>
> dev.off()
null device
1
>
|