Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Prints a measure of uncertainty for 1d function.DescriptionThis function draws the value of a given measure of uncertainty over the whole input domain (1D).
Possible measures are Usageprint_uncertainty_1d(model, T, type = "pn", lower = 0, upper = 1, resolution = 500, new.points = 0, xlab = "", ylab = "", main = "", xscale = c(0, 1), show.points = TRUE, cex.main = 1, cex.lab = 1, cex.points = 1, cex.axis = 1, pch.points.init = 17, pch.points.end = 17, col.points.init = "black", col.points.end = "red", xaxislab = NULL, yaxislab = NULL, xaxispoint = NULL, yaxispoint = NULL, xdecal = 3, ydecal = 3,DiceViewplot=FALSE,vorobmean=FALSE) Arguments
Valuethe integrated uncertainty Author(s)Clement Chevalier (IMSV, Switzerland, and IRSN, France) ReferencesBect J., Ginsbourger D., Li L., Picheny V., Vazquez E. (2010), Sequential design of computer experiments for the estimation of a probability of failure, Statistics and Computing, pp.1-21, 2011, http://arxiv.org/abs/1009.5177 See Also
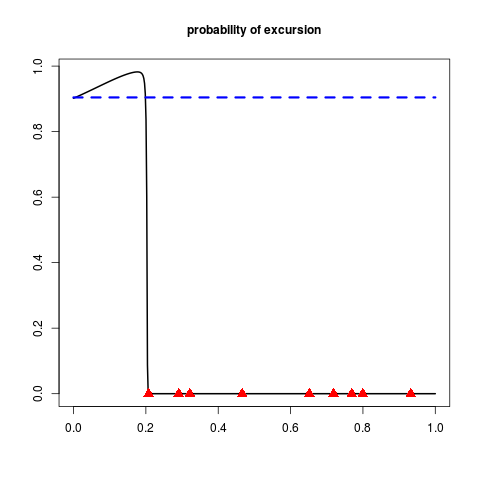
Examples#print_uncertainty_1d set.seed(8) N <- 9 #number of observations T <- 1 #threshold testfun <- fundet lower <- c(0) upper <- c(1) #a 9 points initial design design <- data.frame( matrix(runif(N),ncol=1) ) response <- testfun(design) #km object with matern3_2 covariance #params estimated by ML from the observations model <- km(formula=~., design = design, response = response,covtype="matern3_2") print_uncertainty_1d(DiceViewplot=FALSE,model=model,T=T, main="probability of excursion",cex.points=1.5,col.points.init="red",vorobmean=TRUE) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(KrigInv)
Loading required package: DiceKriging
Loading required package: pbivnorm
Loading required package: rgenoud
## rgenoud (Version 5.7-12.4, Build Date: 2015-07-19)
## See http://sekhon.berkeley.edu/rgenoud for additional documentation.
## Please cite software as:
## Walter Mebane, Jr. and Jasjeet S. Sekhon. 2011.
## ``Genetic Optimization Using Derivatives: The rgenoud package for R.''
## Journal of Statistical Software, 42(11): 1-26.
##
Loading required package: randtoolbox
Loading required package: rngWELL
This is randtoolbox. For overview, type 'help("randtoolbox")'.
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/KrigInv/print_uncertainty_1d.Rd_%03d_medium.png", width=480, height=480)
> ### Name: print_uncertainty_1d
> ### Title: Prints a measure of uncertainty for 1d function.
> ### Aliases: print_uncertainty_1d
>
> ### ** Examples
>
> #print_uncertainty_1d
>
> set.seed(8)
> N <- 9 #number of observations
> T <- 1 #threshold
> testfun <- fundet
> lower <- c(0)
> upper <- c(1)
>
> #a 9 points initial design
> design <- data.frame( matrix(runif(N),ncol=1) )
> response <- testfun(design)
>
> #km object with matern3_2 covariance
> #params estimated by ML from the observations
> model <- km(formula=~., design = design,
+ response = response,covtype="matern3_2")
optimisation start
------------------
* estimation method : MLE
* optimisation method : BFGS
* analytical gradient : used
* trend model : ~matrix.runif.N...ncol...1.
* covariance model :
- type : matern3_2
- nugget : NO
- parameters lower bounds : 1e-10
- parameters upper bounds : 1.448893
- best initial criterion value(s) : 6.967818
N = 1, M = 5 machine precision = 2.22045e-16
At X0, 0 variables are exactly at the bounds
At iterate 0 f= -6.9678 |proj g|= 0.71529
At iterate 1 f = -6.9717 |proj g|= 0.46395
At iterate 2 f = -6.979 |proj g|= 0.077446
At iterate 3 f = -6.9792 |proj g|= 0.010775
At iterate 4 f = -6.9792 |proj g|= 0.0002012
At iterate 5 f = -6.9792 |proj g|= 5.0988e-07
iterations 5
function evaluations 7
segments explored during Cauchy searches 5
BFGS updates skipped 0
active bounds at final generalized Cauchy point 0
norm of the final projected gradient 5.09875e-07
final function value -6.97921
F = -6.97921
final value -6.979214
converged
>
> print_uncertainty_1d(DiceViewplot=FALSE,model=model,T=T,
+ main="probability of excursion",cex.points=1.5,col.points.init="red",vorobmean=TRUE)
[1] 0.1927658
>
>
>
>
>
>
>
> dev.off()
null device
1
>
|