Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
CV for lbDescriptionCross-validation method to tuning the parameter t for lb. Usage
cv.lb(X, y, kappa, alpha, K = 5, tlist, nt = 100, trate = 100,
family = c("gaussian", "binomial", "multinomial"), group = FALSE,
intercept = TRUE, normalize = TRUE, plot.it = TRUE, se = TRUE, ...)
Arguments
DetailsK-fold cross-validation method is used to tuning the parameter t for ISS. Mean square error is used for linear model. Miss-classification error is used for binomial and multinomial model. ValueA list is returned. The list contains a vector of parameter t, crossvalidation error cv.error, and the estimated standard deviation for it cv.sd Author(s)Feng Ruan, Jiechao Xiong and Yuan Yao ReferencesOhser, Ruan, Xiong, Yao and Yin, Sparse Recovery via Differential Inclusions, http://arxiv.org/abs/1406.7728 Examples
#Examples in the reference paper
library(MASS)
n = 200;p = 100;k = 30;sigma = 1
Sigma = 1/(3*p)*matrix(rep(1,p^2),p,p)
diag(Sigma) = 1
A = mvrnorm(n, rep(0, p), Sigma)
u_ref = rep(0,p)
supp_ref = 1:k
u_ref[supp_ref] = rnorm(k)
u_ref[supp_ref] = u_ref[supp_ref]+sign(u_ref[supp_ref])
b = as.vector(A%*%u_ref + sigma*rnorm(n))
cv.lb(A,b,10,1/20,intercept = FALSE,normalize = FALSE)
#Simulated data, binomial case
X <- matrix(rnorm(500*100), nrow=500, ncol=100)
alpha <- c(rep(1,30), rep(0,70))
y <- 2*as.numeric(runif(500)<1/(1+exp(-X %*% alpha)))-1
cv.lb(X,y,kappa=5,alpha=1,family="binomial",
intercept=FALSE,normalize = FALSE)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(Libra)
Loading required package: nnls
Loaded Libra 1.5
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/Libra/cv.lb.Rd_%03d_medium.png", width=480, height=480)
> ### Name: cv.lb
> ### Title: CV for lb
> ### Aliases: cv.lb
> ### Keywords: Cross-validation
>
> ### ** Examples
>
> #Examples in the reference paper
> library(MASS)
> n = 200;p = 100;k = 30;sigma = 1
> Sigma = 1/(3*p)*matrix(rep(1,p^2),p,p)
> diag(Sigma) = 1
> A = mvrnorm(n, rep(0, p), Sigma)
> u_ref = rep(0,p)
> supp_ref = 1:k
> u_ref[supp_ref] = rnorm(k)
> u_ref[supp_ref] = u_ref[supp_ref]+sign(u_ref[supp_ref])
> b = as.vector(A%*%u_ref + sigma*rnorm(n))
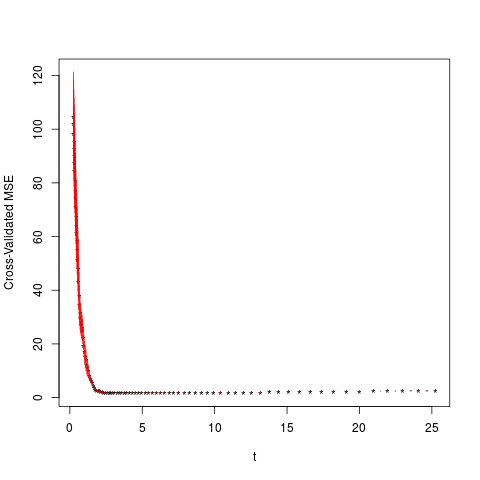
> cv.lb(A,b,10,1/20,intercept = FALSE,normalize = FALSE)
$t
[1] 0.2523626 0.2643791 0.2769677 0.2901557 0.3039717 0.3184455
[7] 0.3336086 0.3494936 0.3661350 0.3835688 0.4018327 0.4209663
[13] 0.4410109 0.4620099 0.4840089 0.5070553 0.5311992 0.5564926
[19] 0.5829904 0.6107500 0.6398313 0.6702973 0.7022140 0.7356505
[25] 0.7706790 0.8073755 0.8458193 0.8860936 0.9282856 0.9724867
[31] 1.0187923 1.0673029 1.1181233 1.1713636 1.2271390 1.2855701
[37] 1.3467835 1.4109116 1.4780932 1.5484738 1.6222055 1.6994481
[43] 1.7803686 1.8651421 1.9539523 2.0469912 2.1444602 2.2465703
[49] 2.3535424 2.4656081 2.5830099 2.7060019 2.8348502 2.9698337
[55] 3.1112446 3.2593889 3.4145871 3.5771752 3.7475051 3.9259454
[61] 4.1128823 4.3087203 4.5138832 4.7288152 4.9539813 5.1898688
[67] 5.4369883 5.6958746 5.9670880 6.2512153 6.5488717 6.8607011
[73] 7.1873786 7.5296110 7.8881391 8.2637388 8.6572229 9.0694431
[79] 9.5012915 9.9537026 10.4276557 10.9241763 11.4443392 11.9892700
[85] 12.5601482 13.1582091 13.7847471 14.4411182 15.1287429 15.8491094
[91] 16.6037767 17.3943780 18.2226244 19.0903084 19.9993078 20.9515899
[97] 21.9492156 22.9943441 24.0892371 25.2362642
$cv.erroe
[1] 104.712736 101.784566 98.052820 95.054531 92.698070 90.083815
[7] 87.284639 84.516695 80.712229 76.982747 74.274930 71.066785
[13] 67.262324 64.005580 61.150352 58.447498 54.879844 51.425946
[19] 47.813554 42.923796 37.894836 34.346578 31.596836 29.624455
[25] 28.200974 26.967204 26.018538 24.679697 22.176077 19.315565
[31] 16.899280 15.303606 13.842156 12.297016 10.955972 9.760491
[37] 8.222686 7.004394 6.041693 5.398641 4.279262 3.493932
[43] 2.998576 2.633723 2.444422 2.315812 2.147801 1.966060
[49] 1.778623 1.673769 1.641720 1.620664 1.617192 1.612369
[55] 1.604392 1.598660 1.595456 1.598252 1.601753 1.637065
[61] 1.658787 1.654317 1.636348 1.636291 1.654309 1.662730
[67] 1.679145 1.690478 1.687726 1.687490 1.684090 1.677448
[73] 1.676405 1.681560 1.697018 1.702904 1.702105 1.728972
[79] 1.749012 1.765516 1.796589 1.868502 1.853434 1.839077
[85] 1.849818 1.886774 1.928899 1.970608 2.002887 2.028296
[91] 2.073326 2.119235 2.168391 2.210397 2.265024 2.375731
[97] 2.433977 2.488238 2.495640 2.505449
$cv.sd
[1] 16.62208543 16.68303859 16.74297442 16.61238403 16.24927925 15.70632680
[7] 15.62608487 15.72565586 14.97825728 14.36985436 14.09277920 13.79473528
[13] 13.56021157 13.32635863 13.01326587 12.63934624 11.93653526 11.77758071
[19] 11.32280419 10.18902898 8.80152025 7.54843591 6.36954414 5.53498542
[25] 5.09188533 4.85683059 4.77107420 4.62049123 4.24865738 4.13405758
[31] 4.18913352 4.27743549 4.04910173 3.56361725 3.03432043 2.45647652
[37] 1.66221988 1.15315178 0.84627660 0.68944615 0.31711013 0.28148847
[43] 0.35281318 0.43189929 0.48558936 0.49568902 0.46114448 0.38929794
[49] 0.25978612 0.19095224 0.16552227 0.15756261 0.15200305 0.14497286
[55] 0.13645817 0.13010530 0.12983105 0.13503011 0.14508433 0.14025339
[61] 0.13897591 0.13703774 0.13371909 0.13693140 0.13555310 0.13582506
[67] 0.13863193 0.14397508 0.14937896 0.15058743 0.13990525 0.13739697
[73] 0.13661281 0.13189917 0.12002795 0.10057141 0.08140745 0.07244043
[79] 0.08677410 0.09919874 0.09991463 0.09797437 0.09958440 0.10063272
[85] 0.09972147 0.10751108 0.12674909 0.12922841 0.12551451 0.11427084
[91] 0.11069609 0.10778147 0.10290944 0.09443625 0.08816807 0.10511478
[97] 0.12589715 0.12887218 0.14410223 0.14684595
>
> #Simulated data, binomial case
> X <- matrix(rnorm(500*100), nrow=500, ncol=100)
> alpha <- c(rep(1,30), rep(0,70))
> y <- 2*as.numeric(runif(500)<1/(1+exp(-X %*% alpha)))-1
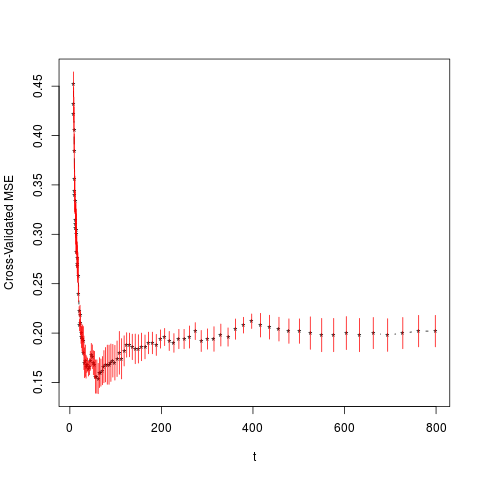
> cv.lb(X,y,kappa=5,alpha=1,family="binomial",
+ intercept=FALSE,normalize = FALSE)
$t
[1] 7.983111 8.363233 8.761455 9.178638 9.615686 10.073544
[7] 10.553203 11.055702 11.582128 12.133619 12.711371 13.316632
[13] 13.950714 14.614987 15.310891 16.039931 16.803684 17.603804
[19] 18.442022 19.320153 20.240097 21.203844 22.213481 23.271193
[25] 24.379268 25.540105 26.756217 28.030234 29.364915 30.763147
[31] 32.227958 33.762516 35.370144 37.054320 38.818689 40.667070
[37] 42.603464 44.632060 46.757249 48.983630 51.316023 53.759474
[43] 56.319272 59.000956 61.810331 64.753477 67.836762 71.066861
[49] 74.450763 77.995792 81.709620 85.600285 89.676207 93.946207
[55] 98.419527 103.105847 108.015309 113.158539 118.546668 124.191357
[61] 130.104822 136.299861 142.789882 149.588929 156.711719 164.173665
[67] 171.990918 180.180395 188.759820 197.747761 207.163670 217.027924
[73] 227.361872 238.187878 249.529373 261.410902 273.858179 286.898143
[79] 300.559014 314.870357 329.863146 345.569829 362.024396 379.262460
[85] 397.321328 416.240082 436.059667 456.822976 478.574946 501.362652
[91] 525.235412 550.244892 576.445217 603.893089 632.647914 662.771920
[97] 694.330304 727.391364 762.026652 798.311124
$cv.erroe
[1] 0.452 0.432 0.422 0.406 0.384 0.356 0.344 0.340 0.334 0.314 0.310 0.306
[13] 0.304 0.300 0.282 0.276 0.270 0.268 0.258 0.240 0.222 0.208 0.218 0.210
[25] 0.200 0.196 0.194 0.194 0.192 0.180 0.170 0.172 0.166 0.168 0.168 0.164
[37] 0.166 0.172 0.178 0.176 0.170 0.168 0.156 0.156 0.154 0.160 0.160 0.162
[49] 0.166 0.168 0.168 0.168 0.170 0.172 0.170 0.174 0.180 0.174 0.182 0.188
[61] 0.188 0.186 0.184 0.184 0.186 0.186 0.190 0.190 0.188 0.194 0.196 0.192
[73] 0.190 0.194 0.194 0.196 0.202 0.192 0.194 0.194 0.198 0.196 0.204 0.208
[85] 0.212 0.208 0.206 0.204 0.202 0.202 0.200 0.198 0.198 0.200 0.198 0.200
[97] 0.198 0.200 0.202 0.202
$cv.sd
[1] 0.012409674 0.016552945 0.016852300 0.022934690 0.017492856 0.016309506
[7] 0.021118712 0.018973666 0.009273618 0.017492856 0.020000000 0.025806976
[13] 0.021587033 0.020736441 0.015937377 0.016309506 0.018708287 0.013564660
[19] 0.018814888 0.008366600 0.004898979 0.005830952 0.010198039 0.011832160
[25] 0.010488088 0.010770330 0.012489996 0.014352700 0.014628739 0.017029386
[31] 0.015165751 0.015937377 0.011661904 0.008000000 0.006633250 0.007483315
[37] 0.008124038 0.009695360 0.011575837 0.012083046 0.012247449 0.013564660
[43] 0.016911535 0.016911535 0.015362291 0.015165751 0.013784049 0.014282857
[49] 0.016309506 0.017435596 0.019849433 0.019849433 0.018973666 0.016552945
[55] 0.017606817 0.018055470 0.021679483 0.020396078 0.015297059 0.012409674
[61] 0.012000000 0.012884099 0.015033296 0.014352700 0.014000000 0.012083046
[67] 0.010954451 0.010954451 0.010677078 0.009273618 0.008717798 0.009695360
[73] 0.009486833 0.009797959 0.009797959 0.011224972 0.008602325 0.010677078
[79] 0.010295630 0.012489996 0.011135529 0.009273618 0.010295630 0.008000000
[85] 0.007348469 0.012000000 0.012083046 0.012083046 0.012409674 0.012409674
[91] 0.016431677 0.016852300 0.016852300 0.016733201 0.016852300 0.015811388
[97] 0.016552945 0.015811388 0.015937377 0.015937377
>
>
>
>
>
>
> dev.off()
null device
1
>
|