Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Pairwise comparison plotDescriptionCreates a heat map from a similarity or Bhattacharyya matrix. UsagepairwisePlot(matrix) Arguments
DetailsThe plot is made using the package ggplot2 and can be reformatted using ggplot2 functions. See examples below. ValueA pairwise comparison heat map. See AlsoAn excellent resource for examples on how to reformat a ggplot can
be found in the R Graphics Cookbook online (http://www.cookbook-r.com/Graphs/).
The functions to create the similarity or Bhattacharyya matrix can be found
here: Examples
file.path <- system.file("extdata", "TCRB_sequencing", package = "LymphoSeq")
file.list <- readImmunoSeq(path = file.path)
productive.aa <- productiveSeq(file.list = file.list, aggregate = "aminoAcid")
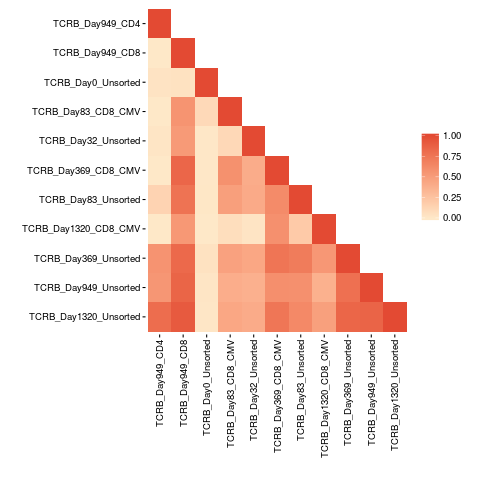
similarity.matrix <- similarityMatrix(productive.seqs = productive.aa)
pairwisePlot(matrix = similarity.matrix)
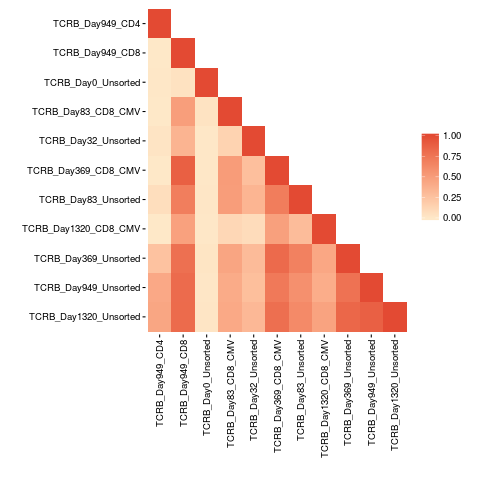
bhattacharyya.matrix <- bhattacharyyaMatrix(productive.seqs = productive.aa)
pairwisePlot(matrix = bhattacharyya.matrix)
# Change plot color, title legend, and add title
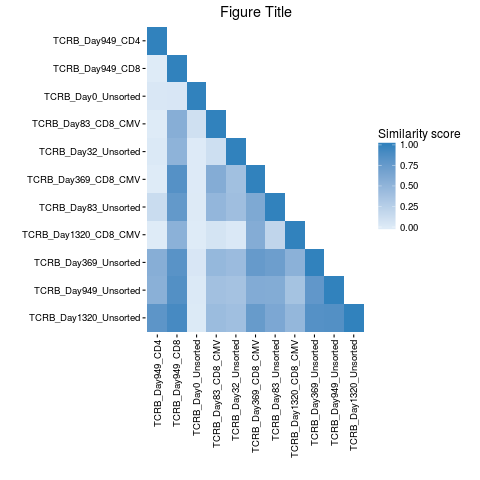
pairwisePlot(matrix = similarity.matrix) +
ggplot2::scale_fill_gradient(low = "#deebf7", high = "#3182bd") +
ggplot2::labs(fill = "Similarity score") + ggplot2::ggtitle("Figure Title")
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(LymphoSeq)
Loading required package: LymphoSeqDB
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/LymphoSeq/pairwisePlot.Rd_%03d_medium.png", width=480, height=480)
> ### Name: pairwisePlot
> ### Title: Pairwise comparison plot
> ### Aliases: pairwisePlot
>
> ### ** Examples
>
> file.path <- system.file("extdata", "TCRB_sequencing", package = "LymphoSeq")
>
> file.list <- readImmunoSeq(path = file.path)
| | | 0% | |====== | 9% | |============= | 18% | |=================== | 27% | |========================= | 36% | |================================ | 45% | |====================================== | 55% | |============================================= | 64% | |=================================================== | 73% | |========================================================= | 82% | |================================================================ | 91% | |======================================================================| 100%
>
> productive.aa <- productiveSeq(file.list = file.list, aggregate = "aminoAcid")
| | | 0% | |====== | 9% | |============= | 18% | |=================== | 27% | |========================= | 36% | |================================ | 45% | |====================================== | 55% | |============================================= | 64% | |=================================================== | 73% | |========================================================= | 82% | |================================================================ | 91% | |======================================================================| 100%
>
> similarity.matrix <- similarityMatrix(productive.seqs = productive.aa)
>
> pairwisePlot(matrix = similarity.matrix)
>
> bhattacharyya.matrix <- bhattacharyyaMatrix(productive.seqs = productive.aa)
>
> pairwisePlot(matrix = bhattacharyya.matrix)
>
> # Change plot color, title legend, and add title
> pairwisePlot(matrix = similarity.matrix) +
+ ggplot2::scale_fill_gradient(low = "#deebf7", high = "#3182bd") +
+ ggplot2::labs(fill = "Similarity score") + ggplot2::ggtitle("Figure Title")
Scale for 'fill' is already present. Adding another scale for 'fill', which
will replace the existing scale.
>
>
>
>
>
> dev.off()
null device
1
>
|