Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Calculation of the melting temperature (Tm) from the first derivativeDescription
UsagediffQ(xy, fct = min, fws = 8, col = 2, plot = FALSE, verbose = FALSE, warn = TRUE, peak = FALSE, negderiv = TRUE, deriv = FALSE, derivlimits = FALSE, derivlimitsline = FALSE, vertiline = FALSE, rsm = FALSE, inder = FALSE) Arguments
Value
Author(s)Stefan Roediger ReferencesA Highly Versatile Microscope Imaging Technology Platform for the Multiplex Real-Time Detection of Biomolecules and Autoimmune Antibodies. S. Roediger, P. Schierack, A. Boehm, J. Nitschke, I. Berger, U. Froemmel, C. Schmidt, M. Ruhland, I. Schimke, D. Roggenbuck, W. Lehmann and C. Schroeder. Advances in Biochemical Bioengineering/Biotechnology. 133:33–74, 2013. http://www.ncbi.nlm.nih.gov/pubmed/22437246 Nucleic acid detection based on the use of microbeads: a review. S. Roediger, C. Liebsch, C. Schmidt, W. Lehmann, U. Resch-Genger, U. Schedler, P. Schierack. Microchim Acta 2014:1–18. DOI: 10.1007/s00604-014-1243-4 Roediger S, Boehm A, Schimke I. Surface Melting Curve Analysis with R. The R Journal 2013;5:37–53. See Also
Examples
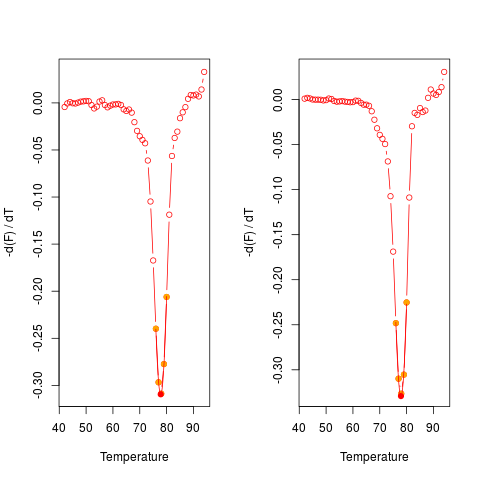
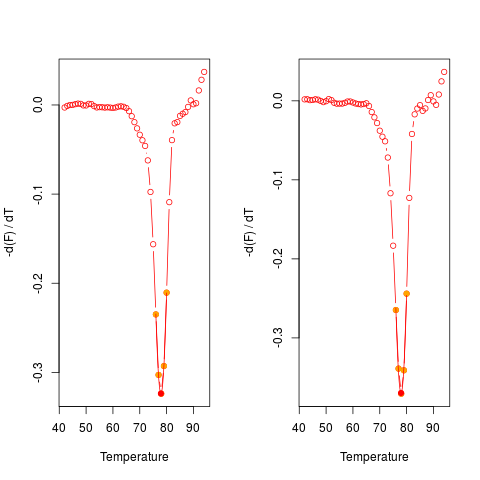
# First Example
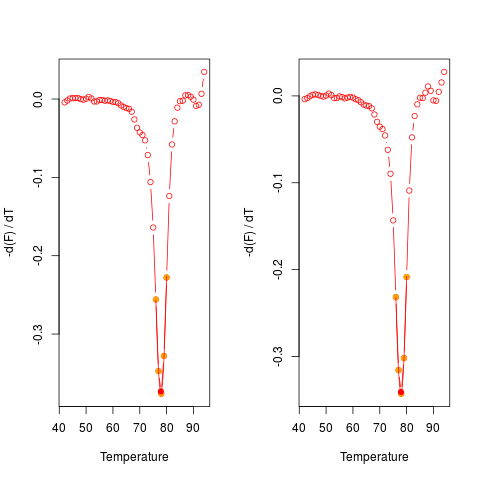
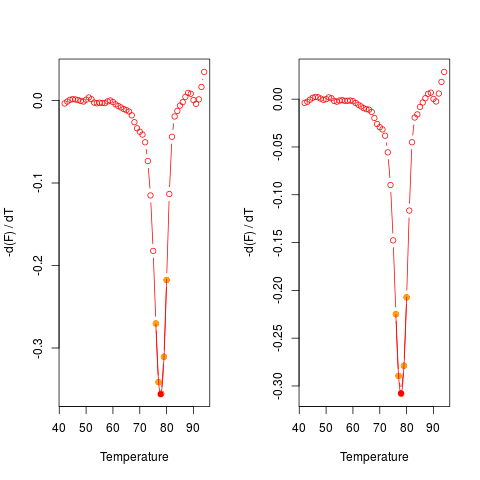
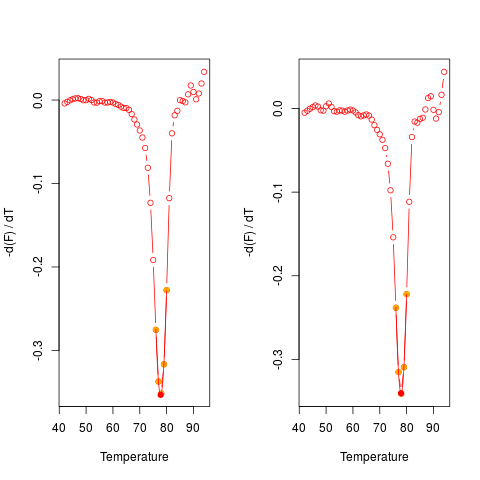
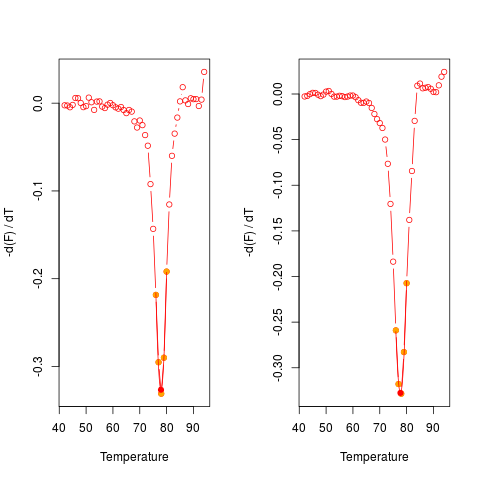
# Plot the first derivative of different samples for single melting curve
# data. Note that the argument "plot" is TRUE.
data(MultiMelt)
par(mfrow = c(1,2))
sapply(2L:14, function(i) {
tmp <- mcaSmoother(MultiMelt[, 1], MultiMelt[, i])
diffQ(tmp, plot = TRUE)
}
)
par(mfrow = c(1,1))
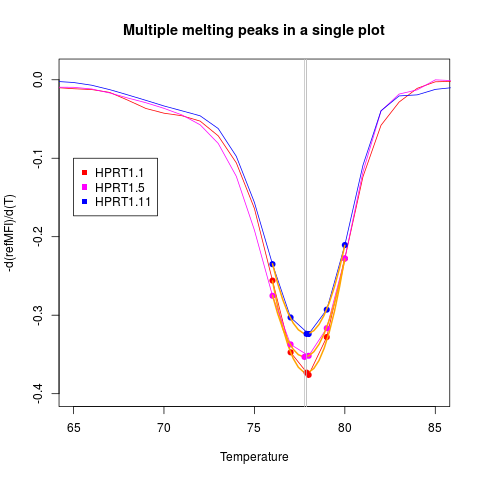
# Second example
# Plot the first derivative of different samples from MultiMelt
# in a single plot.
data(MultiMelt)
# First create an empty plot
plot(NA, NA, xlab = "Temperature", ylab ="-d(refMFI)/d(T)",
main = "Multiple melting peaks in a single plot", xlim = c(65,85),
ylim = c(-0.4,0.01), pch = 19, cex = 1.8)
# Prepossess the selected melting curve data (2,6,12) with mcaSmoother
# and apply them to diffQ. Note that the argument "plot" is FALSE
# while other arguments like derivlimitsline or peak are TRUE.
sapply(c(2,6,12), function(i) {
tmp <- mcaSmoother(MultiMelt[, 1], MultiMelt[, i],
bg = c(41,61), bgadj = TRUE)
diffQ(tmp, plot = FALSE, derivlimitsline = TRUE, deriv = TRUE,
peak = TRUE, derivlimits = TRUE, col = i, vertiline = TRUE)
}
)
legend(65, -0.1, colnames(MultiMelt[, c(2,6,12)]), pch = c(15,15,15),
col = c(2,6,12))
# Third example
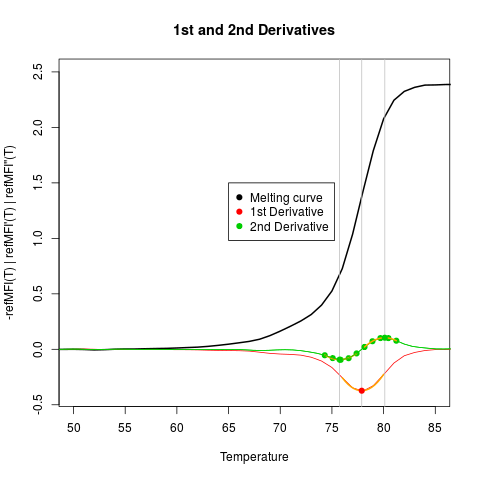
# First create an empty plot
plot(NA, NA, xlim = c(50,85), ylim = c(-0.4,2.5),
xlab = "Temperature",
ylab ="-refMFI(T) | refMFI'(T) | refMFI''(T)",
main = "1st and 2nd Derivatives",
pch = 19, cex = 1.8)
# Prepossess the selected melting curve data with mcaSmoother
# and apply them to diffQ and diffQ2. Note that
# the argument "plot" is FALSE while other
# arguments like derivlimitsline or peak are TRUE.
tmp <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 2],
bg = c(41,61), bgadj = TRUE)
lines(tmp, col= 1, lwd = 2)
# Note the different use of the argument derivlimits in diffQ and diffQ2
diffQ(tmp, fct = min, derivlimitsline = TRUE, deriv = TRUE,
peak = TRUE, derivlimits = FALSE, col = 2, vertiline = TRUE)
diffQ2(tmp, fct = min, derivlimitsline = TRUE, deriv = TRUE,
peak = TRUE, derivlimits = TRUE, col = 3, vertiline = TRUE)
# Add a legend to the plot
legend(65, 1.5, c("Melting curve",
"1st Derivative",
"2nd Derivative"),
pch = c(19,19,19), col = c(1,2,3))
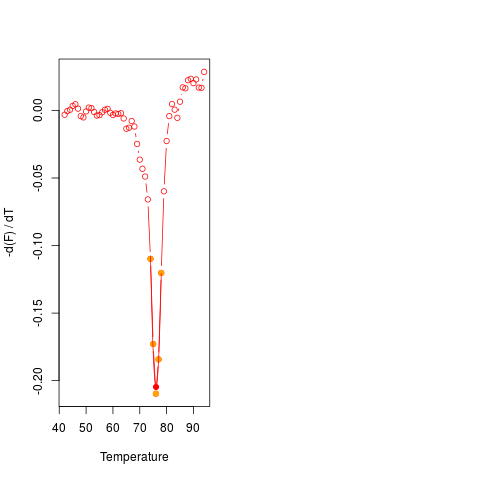
# Fourth example
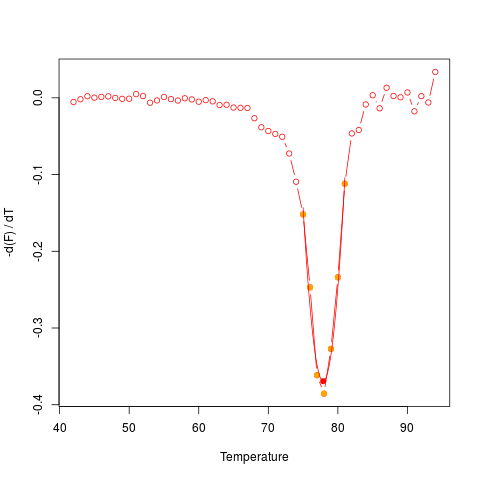
# Different curves may potentially have different quality in practice.
# For example, using data from MultiMelt should return a
# valid result and plot.
data(MultiMelt)
diffQ(cbind(MultiMelt[, 1], MultiMelt[, 2]), plot = TRUE)$Tm
# limits_xQ
# 77.88139
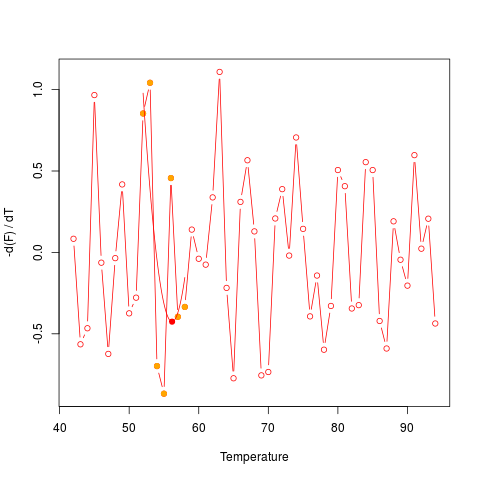
# Imagine an experiment that went terribly wrong. You would
# still get an estimate for the Tm. The output from diffQ,
# with an error attached, lets the user know that this Tm
# is potentially meaningless. diffQ() will give several
# warning messages.
set.seed(1)
y = rnorm(55,1.5,.8)
diffQ(cbind(MultiMelt[, 1],y), plot = TRUE)$Tm
# The distribution of the curve data indicates noise.
# The data should be visually inspected with a plot
# (see examples of diffQ). The Tm calculation (fit,
# adj. R squared ~ 0.157, NRMSE ~ 0.279) is not optimal
# presumably due to noisy data. Check raw melting
# curve (see examples of diffQ).
# Calculated Tm
# 56.16755
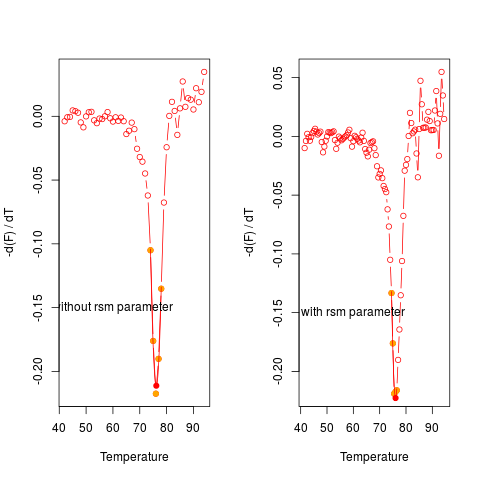
# Sixth example
# Curves may potentially have a low temperature resolution. The rsm
# parameter can be used to double the temperature resolution.
# Use data from MultiMelt column 15 (MLC2v2).
data(MultiMelt)
tmp <- cbind(MultiMelt[, 1], MultiMelt[, 15])
# Use diffQ without and with the rsm parameter and plot
# the results in a single row
par(mfrow = c(1,2))
diffQ(tmp, plot = TRUE)$Tm
text(60, -0.15, "without rsm parameter")
diffQ(tmp, plot = TRUE, rsm = TRUE)$Tm
text(60, -0.15, "with rsm parameter")
par(mfrow = c(1,1))
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(MBmca)
Loading required package: robustbase
Loading required package: chipPCR
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/MBmca/diffQ.Rd_%03d_medium.png", width=480, height=480)
> ### Name: diffQ
> ### Title: Calculation of the melting temperature (Tm) from the first
> ### derivative
> ### Aliases: diffQ
> ### Keywords: Tm melting
>
> ### ** Examples
>
> # First Example
> # Plot the first derivative of different samples for single melting curve
> # data. Note that the argument "plot" is TRUE.
>
> data(MultiMelt)
> par(mfrow = c(1,2))
> sapply(2L:14, function(i) {
+ tmp <- mcaSmoother(MultiMelt[, 1], MultiMelt[, i])
+ diffQ(tmp, plot = TRUE)
+ }
+ )
[,1] [,2] [,3] [,4] [,5] [,6]
Tm 77.88494 77.90029 77.75343 77.89949 77.76936 77.93054
fluoTm -0.3732338 -0.3408576 -0.3560144 -0.3076871 -0.3532675 -0.3405116
[,7] [,8] [,9] [,10] [,11] [,12]
Tm 77.90438 77.7017 77.79736 77.88993 77.8833 77.93199
fluoTm -0.3265402 -0.3275499 -0.3095232 -0.3292306 -0.3237404 -0.369646
[,13]
Tm 76.07117
fluoTm -0.2046684
> par(mfrow = c(1,1))
> # Second example
> # Plot the first derivative of different samples from MultiMelt
> # in a single plot.
> data(MultiMelt)
>
> # First create an empty plot
> plot(NA, NA, xlab = "Temperature", ylab ="-d(refMFI)/d(T)",
+ main = "Multiple melting peaks in a single plot", xlim = c(65,85),
+ ylim = c(-0.4,0.01), pch = 19, cex = 1.8)
> # Prepossess the selected melting curve data (2,6,12) with mcaSmoother
> # and apply them to diffQ. Note that the argument "plot" is FALSE
> # while other arguments like derivlimitsline or peak are TRUE.
> sapply(c(2,6,12), function(i) {
+ tmp <- mcaSmoother(MultiMelt[, 1], MultiMelt[, i],
+ bg = c(41,61), bgadj = TRUE)
+ diffQ(tmp, plot = FALSE, derivlimitsline = TRUE, deriv = TRUE,
+ peak = TRUE, derivlimits = TRUE, col = i, vertiline = TRUE)
+ }
+ )
[,1] [,2] [,3]
Tm 77.88494 77.76936 77.8833
fluoTm -0.3731492 -0.3532051 -0.3237496
> legend(65, -0.1, colnames(MultiMelt[, c(2,6,12)]), pch = c(15,15,15),
+ col = c(2,6,12))
>
> # Third example
> # First create an empty plot
> plot(NA, NA, xlim = c(50,85), ylim = c(-0.4,2.5),
+ xlab = "Temperature",
+ ylab ="-refMFI(T) | refMFI'(T) | refMFI''(T)",
+ main = "1st and 2nd Derivatives",
+ pch = 19, cex = 1.8)
>
> # Prepossess the selected melting curve data with mcaSmoother
> # and apply them to diffQ and diffQ2. Note that
> # the argument "plot" is FALSE while other
> # arguments like derivlimitsline or peak are TRUE.
>
> tmp <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 2],
+ bg = c(41,61), bgadj = TRUE)
> lines(tmp, col= 1, lwd = 2)
>
> # Note the different use of the argument derivlimits in diffQ and diffQ2
> diffQ(tmp, fct = min, derivlimitsline = TRUE, deriv = TRUE,
+ peak = TRUE, derivlimits = FALSE, col = 2, vertiline = TRUE)
Calculated Tm: 77.88494
Signal height at calculated Tm: -0.3731492
> diffQ2(tmp, fct = min, derivlimitsline = TRUE, deriv = TRUE,
+ peak = TRUE, derivlimits = TRUE, col = 3, vertiline = TRUE)
Calculated Tm: 77.88494
Signal height at calculated Tm: -0.3731492
Calculated 'left' Tm: 75.74357
Calculated 'left' signal height: -0.0931392
Calculated 'right' Tm: 80.11672
Calculated 'right' signal height: 0.1042746
>
> # Add a legend to the plot
> legend(65, 1.5, c("Melting curve",
+ "1st Derivative",
+ "2nd Derivative"),
+ pch = c(19,19,19), col = c(1,2,3))
>
> # Fourth example
> # Different curves may potentially have different quality in practice.
> # For example, using data from MultiMelt should return a
> # valid result and plot.
> data(MultiMelt)
>
> diffQ(cbind(MultiMelt[, 1], MultiMelt[, 2]), plot = TRUE)$Tm
Calculated Tm
77.88139
> # limits_xQ
> # 77.88139
>
> # Imagine an experiment that went terribly wrong. You would
> # still get an estimate for the Tm. The output from diffQ,
> # with an error attached, lets the user know that this Tm
> # is potentially meaningless. diffQ() will give several
> # warning messages.
>
> set.seed(1)
> y = rnorm(55,1.5,.8)
> diffQ(cbind(MultiMelt[, 1],y), plot = TRUE)$Tm
The distribution of the curve data indicates noise. The data should be visually
inspected with a plot (see examples of diffQ).
The Tm calculation (fit, adj. R squared ~ 0.157, NRMSE ~ 0.279) is not optimal presumably
due to noisy data. Check raw melting curve (see examples of diffQ).
Calculated Tm
56.16755
>
> # The distribution of the curve data indicates noise.
> # The data should be visually inspected with a plot
> # (see examples of diffQ). The Tm calculation (fit,
> # adj. R squared ~ 0.157, NRMSE ~ 0.279) is not optimal
> # presumably due to noisy data. Check raw melting
> # curve (see examples of diffQ).
> # Calculated Tm
> # 56.16755
>
>
> # Sixth example
> # Curves may potentially have a low temperature resolution. The rsm
> # parameter can be used to double the temperature resolution.
> # Use data from MultiMelt column 15 (MLC2v2).
> data(MultiMelt)
> tmp <- cbind(MultiMelt[, 1], MultiMelt[, 15])
>
> # Use diffQ without and with the rsm parameter and plot
> # the results in a single row
> par(mfrow = c(1,2))
>
> diffQ(tmp, plot = TRUE)$Tm
Calculated Tm
76.16234
> text(60, -0.15, "without rsm parameter")
>
> diffQ(tmp, plot = TRUE, rsm = TRUE)$Tm
Calculated Tm
76.04511
> text(60, -0.15, "with rsm parameter")
> par(mfrow = c(1,1))
>
>
>
>
>
> dev.off()
null device
1
>
|