Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Function to pre-process melting curve data.DescriptionThe function UsagemcaSmoother(x, y, bgadj = FALSE, bg = NULL, Trange = NULL, minmax = FALSE, df.fact = 0.95, n = NULL) Arguments
Value
Author(s)Stefan Roediger ReferencesA Highly Versatile Microscope Imaging Technology Platform for the Multiplex Real-Time Detection of Biomolecules and Autoimmune Antibodies. S. Roediger, P. Schierack, A. Boehm, J. Nitschke, I. Berger, U. Froemmel, C. Schmidt, M. Ruhland, I. Schimke, D. Roggenbuck, W. Lehmann and C. Schroeder. Advances in Biochemical Bioengineering/Biotechnology. 133:33–74, 2013. http://www.ncbi.nlm.nih.gov/pubmed/22437246 Nucleic acid detection based on the use of microbeads: a review. S. Roediger, C. Liebsch, C. Schmidt, W. Lehmann, U. Resch-Genger, U. Schedler, P. Schierack. Microchim Acta 2014:1–18. DOI: 10.1007/s00604-014-1243-4 Roediger S, Boehm A, Schimke I. Surface Melting Curve Analysis with R. The R Journal 2013;5:37–53. See Also
Examples
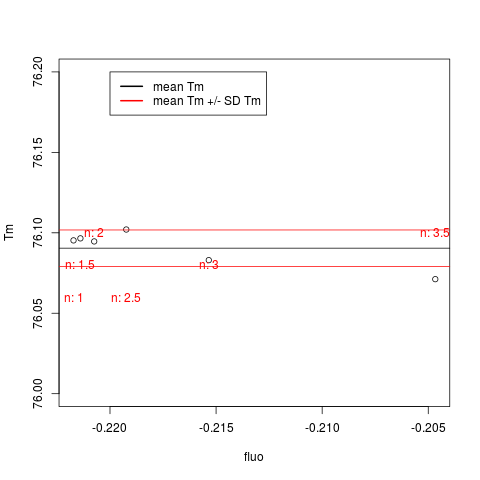
# First Example
# Use mcaSmoother with different n to increase the temperature
# resolution of the melting curve artificially. Compare the
# influence of the n on the Tm and fluoTm values
data(MultiMelt)
Tm <- vector()
fluo <- vector()
for (i in seq(1,3.5,0.5)) {
res.smooth <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 14], n = i)
res <- diffQ(res.smooth)
Tm <- c(Tm, res$Tm)
fluo <- c(fluo, res$fluoTm)
}
plot(fluo, Tm, ylim = c(76,76.2))
abline(h = mean(Tm))
text(fluo, seq(76.1,76.05,-0.02),
paste("n:", seq(3.5,1,-0.5), sep = " "), col = 2)
abline(h = c(mean(Tm) + sd(Tm), mean(Tm) - sd(Tm)), col = 2)
legend(-0.22, 76.2, c("mean Tm", "mean Tm +/- SD Tm"),
col = c(1,2), lwd = 2)
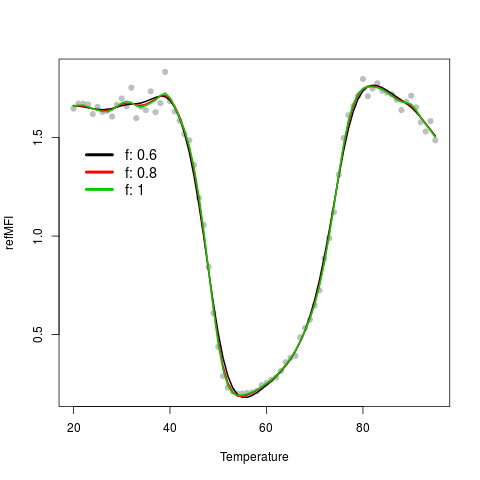
# Second Example
# Use mcaSmoother with different strengths of smoothing
# (f, 0.6 = strongest, 1 = weakest).
data(DMP)
plot(DMP[, 1], DMP[,6],
xlim = c(20,95), xlab = "Temperature",
ylab = "refMFI", pch = 19, col = 8)
f <- c(0.6, 0.8, 1.0)
for (i in c(1:3)) {
lines(mcaSmoother(DMP[, 1],
DMP[,6], df.fact = f[i]),
col = i, lwd = 2)
}
legend(20, 1.5, paste("f", f, sep = ": "),
cex = 1.2, col = 1:3, bty = "n",
lty = 1, lwd = 4)
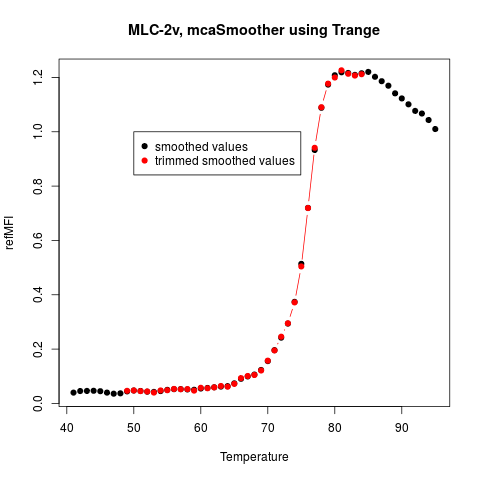
# Third Example
# Plot the smoothed and trimmed melting curve
data(MultiMelt)
tmp <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 14])
tmp.trimmed <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 14],
Trange = c(49,85))
plot(tmp, pch = 19, xlab = "Temperature", ylab = "refMFI",
main = "MLC-2v, mcaSmoother using Trange")
points(tmp.trimmed, col = 2, type = "b", pch = 19)
legend(50, 1, c("smoothed values",
"trimmed smoothed values"),
pch = c(19,19), col = c(1,2))
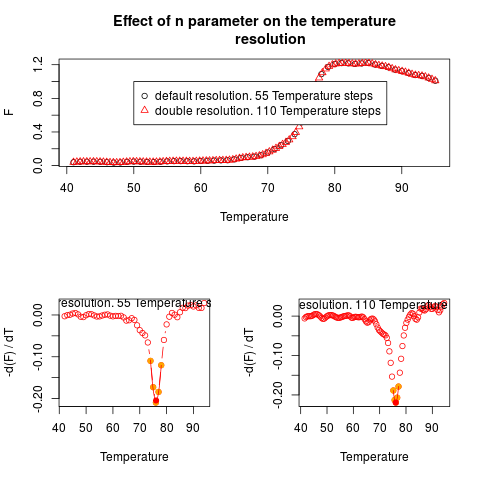
# Fourth Example
# Use mcaSmoother with different n to increase the temperature
# resolution of the melting curve. Caution, this operation may
# affect your data negatively if the resolution is set to high.
# Higher resolutions will just give the impression of better
# data quality. res.st uses the default resolution (no
# alteration)
# res.high uses the double resolution.
data(MultiMelt)
res.st <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 14])
res.high <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 14], n = 2)
par(fig = c(0,1,0.5,1))
plot(res.st, xlab = "Temperature", ylab = "F",
main = "Effect of n parameter on the temperature
resolution")
points(res.high, col = 2, pch = 2)
legend(50, 1, c(paste("default resolution.", nrow(res.st),
"Temperature steps", sep = " "),
paste("double resolution.", nrow(res.high),
"Temperature steps", sep = " ")),
pch = c(1,2), col = c(1,2))
par(fig = c(0,0.5,0,0.5), new = TRUE)
diffQ(res.st, plot = TRUE)
text(65, 0.025, paste("default resolution.", nrow(res.st),
"Temperature steps", sep = " "))
par(fig = c(0.5,1,0,0.5), new = TRUE)
diffQ(res.high, plot = TRUE)
text(65, 0.025, paste("double resolution.", nrow(res.high),
"Temperature steps", sep = " "))
# Fifth example
# Different experiments may have different temperature
# resolutions and temperature ranges. The example uses a
# simulated melting curve with a temperature resolution of
# 0.5 and 1 degree Celsius and a temperature range of
# 35 to 95 degree Celsius.
#
# Coefficients of a 3 parameter sigmoid model. Note:
# The off-set, temperature range and temperature resolution
# differ between both simulations. However, the melting
# temperatures should be very
# similar finally.
b <- -0.5; e <- 77
# Simulate first melting curve with a temperature
# between 35 - 95 degree Celsius and 1 degree Celsius
# per step temperature resolution.
t1 <- seq(35, 95, 1)
f1 <- 0.3 + 4 / (1 + exp(b * (t1 - e)))
# Simulate second melting curve with a temperature
# between 41.5 - 92.1 degree Celsius and 0.5 degree Celsius
# per step temperature resolution.
t2 <- seq(41.5, 92.1, 0.5)
f2 <- 0.2 + 2 / (1 + exp(b * (t2 - e)))
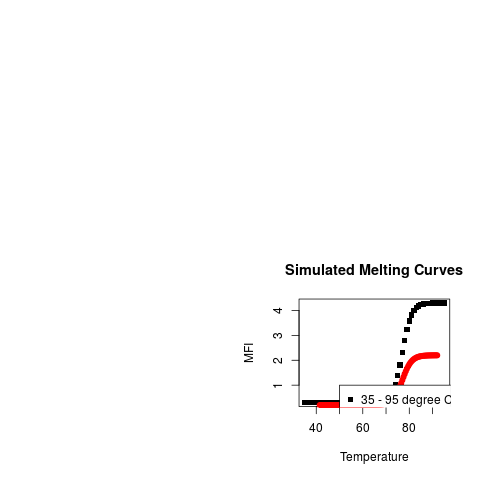
# Plot both simulated melting curves
plot(t1, f1, pch = 15, ylab = "MFI",
main = "Simulated Melting Curves",
xlab = "Temperature", col = 1)
points(t2, f2, pch = 19, col = 2)
legend(50, 1,
c("35 - 95 degree Celsius, 1 degree Celsius per step",
"41.5 - 92.1 degree Celsius, 0.5 degree Celsius per step",
sep = " "), pch = c(15,19), col = c(1,2))
# Use mcaSmoother with n = 2 to increase the temperature
# resolution of the first simulated melting curve. The minmax
# parameter is used to make the peak heights compareable. The
# temperature range was limited between 45 to 90 degree Celsius for
# both simulations
t1f1 <- mcaSmoother(t1, f1, Trange= c(45, 90), minmax = TRUE, n = 2)
t2f2 <- mcaSmoother(t2, f2, Trange= c(45, 90), minmax = TRUE, n = 1)
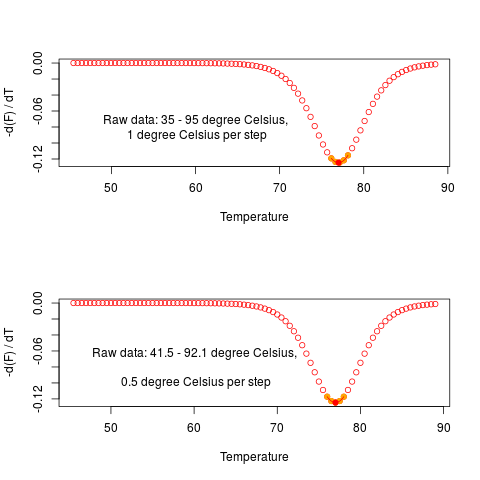
# Perform a MCA on both altered simulations. As expected, the melting
# temperature are almost identical.
par(mfrow = c(2,1))
# Tm 77.00263, fluoTm -0.1245848
diffQ(t1f1, plot = TRUE)
text(60, -0.08,
"Raw data: 35 - 95 degree Celsius,\n 1 degree Celsius per step")
# Tm 77.00069, fluoTm -0.1245394
diffQ(t2f2, plot = TRUE)
text(60, -0.08, "Raw data: 41.5 - 92.1 degree Celsius,
\n 0.5 degree Celsius per step")
par(mfrow = c(1,1))
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(MBmca)
Loading required package: robustbase
Loading required package: chipPCR
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/MBmca/mcaSmoother.Rd_%03d_medium.png", width=480, height=480)
> ### Name: mcaSmoother
> ### Title: Function to pre-process melting curve data.
> ### Aliases: mcaSmoother
> ### Keywords: background normalization smooth
>
> ### ** Examples
>
> # First Example
> # Use mcaSmoother with different n to increase the temperature
> # resolution of the melting curve artificially. Compare the
> # influence of the n on the Tm and fluoTm values
> data(MultiMelt)
>
> Tm <- vector()
> fluo <- vector()
> for (i in seq(1,3.5,0.5)) {
+ res.smooth <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 14], n = i)
+ res <- diffQ(res.smooth)
+ Tm <- c(Tm, res$Tm)
+ fluo <- c(fluo, res$fluoTm)
+ }
> plot(fluo, Tm, ylim = c(76,76.2))
> abline(h = mean(Tm))
> text(fluo, seq(76.1,76.05,-0.02),
+ paste("n:", seq(3.5,1,-0.5), sep = " "), col = 2)
> abline(h = c(mean(Tm) + sd(Tm), mean(Tm) - sd(Tm)), col = 2)
>
> legend(-0.22, 76.2, c("mean Tm", "mean Tm +/- SD Tm"),
+ col = c(1,2), lwd = 2)
>
> # Second Example
> # Use mcaSmoother with different strengths of smoothing
> # (f, 0.6 = strongest, 1 = weakest).
> data(DMP)
> plot(DMP[, 1], DMP[,6],
+ xlim = c(20,95), xlab = "Temperature",
+ ylab = "refMFI", pch = 19, col = 8)
> f <- c(0.6, 0.8, 1.0)
> for (i in c(1:3)) {
+ lines(mcaSmoother(DMP[, 1],
+ DMP[,6], df.fact = f[i]),
+ col = i, lwd = 2)
+ }
> legend(20, 1.5, paste("f", f, sep = ": "),
+ cex = 1.2, col = 1:3, bty = "n",
+ lty = 1, lwd = 4)
>
> # Third Example
> # Plot the smoothed and trimmed melting curve
> data(MultiMelt)
> tmp <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 14])
> tmp.trimmed <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 14],
+ Trange = c(49,85))
> plot(tmp, pch = 19, xlab = "Temperature", ylab = "refMFI",
+ main = "MLC-2v, mcaSmoother using Trange")
> points(tmp.trimmed, col = 2, type = "b", pch = 19)
> legend(50, 1, c("smoothed values",
+ "trimmed smoothed values"),
+ pch = c(19,19), col = c(1,2))
>
> # Fourth Example
> # Use mcaSmoother with different n to increase the temperature
> # resolution of the melting curve. Caution, this operation may
> # affect your data negatively if the resolution is set to high.
> # Higher resolutions will just give the impression of better
> # data quality. res.st uses the default resolution (no
> # alteration)
> # res.high uses the double resolution.
> data(MultiMelt)
> res.st <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 14])
> res.high <- mcaSmoother(MultiMelt[, 1], MultiMelt[, 14], n = 2)
>
> par(fig = c(0,1,0.5,1))
> plot(res.st, xlab = "Temperature", ylab = "F",
+ main = "Effect of n parameter on the temperature
+ resolution")
> points(res.high, col = 2, pch = 2)
> legend(50, 1, c(paste("default resolution.", nrow(res.st),
+ "Temperature steps", sep = " "),
+ paste("double resolution.", nrow(res.high),
+ "Temperature steps", sep = " ")),
+ pch = c(1,2), col = c(1,2))
> par(fig = c(0,0.5,0,0.5), new = TRUE)
> diffQ(res.st, plot = TRUE)
Calculated Tm: 76.07117
Signal height at calculated Tm: -0.2046684
> text(65, 0.025, paste("default resolution.", nrow(res.st),
+ "Temperature steps", sep = " "))
> par(fig = c(0.5,1,0,0.5), new = TRUE)
> diffQ(res.high, plot = TRUE)
Calculated Tm: 76.10204
Signal height at calculated Tm: -0.2192319
> text(65, 0.025, paste("double resolution.", nrow(res.high),
+ "Temperature steps", sep = " "))
>
> # Fifth example
> # Different experiments may have different temperature
> # resolutions and temperature ranges. The example uses a
> # simulated melting curve with a temperature resolution of
> # 0.5 and 1 degree Celsius and a temperature range of
> # 35 to 95 degree Celsius.
> #
> # Coefficients of a 3 parameter sigmoid model. Note:
> # The off-set, temperature range and temperature resolution
> # differ between both simulations. However, the melting
> # temperatures should be very
> # similar finally.
> b <- -0.5; e <- 77
>
> # Simulate first melting curve with a temperature
> # between 35 - 95 degree Celsius and 1 degree Celsius
> # per step temperature resolution.
>
> t1 <- seq(35, 95, 1)
> f1 <- 0.3 + 4 / (1 + exp(b * (t1 - e)))
>
> # Simulate second melting curve with a temperature
> # between 41.5 - 92.1 degree Celsius and 0.5 degree Celsius
> # per step temperature resolution.
> t2 <- seq(41.5, 92.1, 0.5)
> f2 <- 0.2 + 2 / (1 + exp(b * (t2 - e)))
>
> # Plot both simulated melting curves
> plot(t1, f1, pch = 15, ylab = "MFI",
+ main = "Simulated Melting Curves",
+ xlab = "Temperature", col = 1)
> points(t2, f2, pch = 19, col = 2)
> legend(50, 1,
+ c("35 - 95 degree Celsius, 1 degree Celsius per step",
+ "41.5 - 92.1 degree Celsius, 0.5 degree Celsius per step",
+ sep = " "), pch = c(15,19), col = c(1,2))
>
> # Use mcaSmoother with n = 2 to increase the temperature
> # resolution of the first simulated melting curve. The minmax
> # parameter is used to make the peak heights compareable. The
> # temperature range was limited between 45 to 90 degree Celsius for
> # both simulations
>
> t1f1 <- mcaSmoother(t1, f1, Trange= c(45, 90), minmax = TRUE, n = 2)
> t2f2 <- mcaSmoother(t2, f2, Trange= c(45, 90), minmax = TRUE, n = 1)
>
> # Perform a MCA on both altered simulations. As expected, the melting
> # temperature are almost identical.
> par(mfrow = c(2,1))
> # Tm 77.00263, fluoTm -0.1245848
> diffQ(t1f1, plot = TRUE)
Calculated Tm: 77.00263
Signal height at calculated Tm: -0.1245848
> text(60, -0.08,
+ "Raw data: 35 - 95 degree Celsius,\n 1 degree Celsius per step")
>
> # Tm 77.00069, fluoTm -0.1245394
> diffQ(t2f2, plot = TRUE)
Calculated Tm: 77.00069
Signal height at calculated Tm: -0.1245394
> text(60, -0.08, "Raw data: 41.5 - 92.1 degree Celsius,
+ \n 0.5 degree Celsius per step")
> par(mfrow = c(1,1))
>
>
>
>
>
> dev.off()
null device
1
>
|