Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Markov Chain Monte Carlo for Ordered Probit Changepoint Regression ModelDescriptionThis function generates a sample from the posterior distribution of an ordered probit regression model with multiple parameter breaks. The function uses the Markov chain Monte Carlo method of Chib (1998). The user supplies data and priors, and a sample from the posterior distribution is returned as an mcmc object, which can be subsequently analyzed with functions provided in the coda package. UsageMCMCoprobitChange(formula, data=parent.frame(), m = 1,
burnin = 1000, mcmc = 1000, thin = 1, tune = NA, verbose = 0,
seed = NA, beta.start = NA, gamma.start=NA, P.start = NA,
b0 = NULL, B0 = NULL, a = NULL, b = NULL,
marginal.likelihood = c("none", "Chib95"), gamma.fixed=0, ...)
Arguments
Details
The model takes the following form: Pr(y_t = 1) = Phi(gamma_(c, m) - x_i'beta_m) - Phi(gamma_(c-1, m) - x_i'beta) Where M is the number of states, and gamma_(c, m) and beta_m are paramters when a state is m at t. We assume Gaussian distribution for prior of beta: beta_m ~ N(b0,B0^(-1)), m = 1,...,M. And: p_mm ~ Beta(a, b), m = 1,...,M. Where M is the number of states. Note that when the fitted changepoint model has very few observations in any of states, the marginal likelihood outcome can be “nan," which indicates that too many breaks are assumed given the model and data. ValueAn mcmc object that contains the posterior sample. This
object can be summarized by functions provided by the coda package.
The object contains an attribute ReferencesJong Hee Park. 2011. “Changepoint Analysis of Binary and Ordinal Probit Models: An Application to Bank Rate Policy Under the Interwar Gold Standard." Political Analysis. 19: 188-204. Andrew D. Martin, Kevin M. Quinn, and Jong Hee Park. 2011. “MCMCpack: Markov Chain Monte Carlo in R.”, Journal of Statistical Software. 42(9): 1-21. http://www.jstatsoft.org/v42/i09/. Siddhartha Chib. 1998. “Estimation and comparison of multiple change-point models.” Journal of Econometrics. 86: 221-241. See Also
Examples
set.seed(1909)
N <- 200
x1 <- rnorm(N, 1, .5);
## set a true break at 100
z1 <- 1 + x1[1:100] + rnorm(100);
z2 <- 1 -0.2*x1[101:200] + rnorm(100);
z <- c(z1, z2);
y <- z
## generate y
y[z < 1] <- 1;
y[z >= 1 & z < 2] <- 2;
y[z >= 2] <- 3;
## inputs
formula <- y ~ x1
## fit multiple models with a varying number of breaks
out1 <- MCMCoprobitChange(formula, m=1,
mcmc=100, burnin=100, thin=1, tune=c(.5, .5), verbose=100,
b0=0, B0=10, marginal.likelihood = "Chib95")
out2 <- MCMCoprobitChange(formula, m=2,
mcmc=100, burnin=100, thin=1, tune=c(.5, .5, .5), verbose=100,
b0=0, B0=10, marginal.likelihood = "Chib95")
## Do model comparison
## NOTE: the chain should be run longer than this example!
BayesFactor(out1, out2)
## draw plots using the "right" model
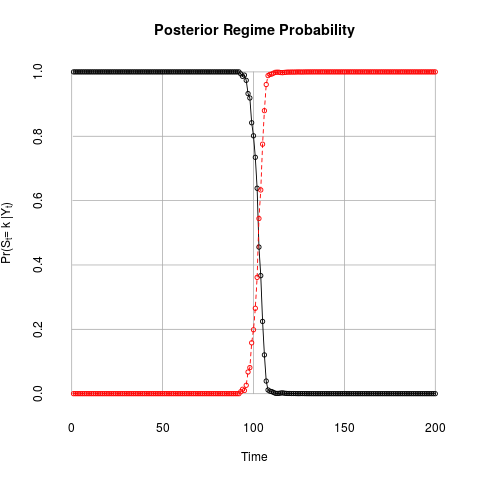
plotState(out1)
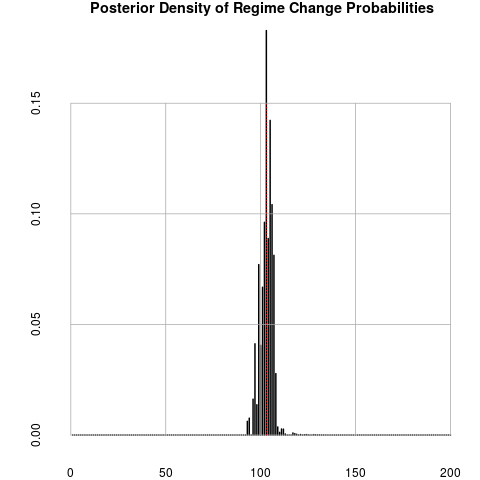
plotChangepoint(out1)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(MCMCpack)
Loading required package: coda
Loading required package: MASS
##
## Markov Chain Monte Carlo Package (MCMCpack)
## Copyright (C) 2003-2016 Andrew D. Martin, Kevin M. Quinn, and Jong Hee Park
##
## Support provided by the U.S. National Science Foundation
## (Grants SES-0350646 and SES-0350613)
##
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/MCMCpack/MCMCoprobitChange.Rd_%03d_medium.png", width=480, height=480)
> ### Name: MCMCoprobitChange
> ### Title: Markov Chain Monte Carlo for Ordered Probit Changepoint
> ### Regression Model
> ### Aliases: MCMCoprobitChange
> ### Keywords: models
>
> ### ** Examples
>
> set.seed(1909)
> N <- 200
> x1 <- rnorm(N, 1, .5);
>
> ## set a true break at 100
> z1 <- 1 + x1[1:100] + rnorm(100);
> z2 <- 1 -0.2*x1[101:200] + rnorm(100);
> z <- c(z1, z2);
> y <- z
>
> ## generate y
> y[z < 1] <- 1;
> y[z >= 1 & z < 2] <- 2;
> y[z >= 2] <- 3;
>
> ## inputs
> formula <- y ~ x1
>
> ## fit multiple models with a varying number of breaks
> out1 <- MCMCoprobitChange(formula, m=1,
+ mcmc=100, burnin=100, thin=1, tune=c(.5, .5), verbose=100,
+ b0=0, B0=10, marginal.likelihood = "Chib95")
MCMCoprobitChange iteration 101 of 200
Acceptance rate for state 1 is 0.53000
Acceptance rate for state 2 is 0.20000
The number of observations in state 1 is 00097
The number of observations in state 2 is 00103
beta 0 = 0.09304 0.52479
beta 1 = -0.27336 0.16873
gamma 0 = 0.74271
gamma 1 = 0.99581
logmarglike = -220.60851
loglike = -192.32574
log_prior = -4.61781
log_beta = 4.33846
log_P = 3.63139
log_gamma = 15.69512
> out2 <- MCMCoprobitChange(formula, m=2,
+ mcmc=100, burnin=100, thin=1, tune=c(.5, .5, .5), verbose=100,
+ b0=0, B0=10, marginal.likelihood = "Chib95")
MCMCoprobitChange iteration 101 of 200
Acceptance rate for state 1 is 0.61000
Acceptance rate for state 2 is 0.81000
Acceptance rate for state 3 is 0.21000
The number of observations in state 1 is 00001
The number of observations in state 2 is 00001
The number of observations in state 3 is 00198
beta 0 = 0.09056 0.25382
beta 1 = -0.54118 -0.45300
beta 2 = -0.33464 0.54185
gamma 0 = 0.92141
gamma 1 = 1.23897
gamma 2 = 0.70838
logmarglike = -241.98699
loglike = -212.84326
log_prior = -8.97530
log_beta = 3.75571
log_P = 2.18454
log_gamma = 14.22818
>
> ## Do model comparison
> ## NOTE: the chain should be run longer than this example!
> BayesFactor(out1, out2)
The matrix of Bayes Factors is:
out1 out2
out1 1.00e+00 1.93e+09
out2 5.19e-10 1.00e+00
The matrix of the natural log Bayes Factors is:
out1 out2
out1 0.0 21.4
out2 -21.4 0.0
out1 :
call =
MCMCoprobitChange(formula = formula, m = 1, burnin = 100, mcmc = 100,
thin = 1, tune = c(0.5, 0.5), verbose = 100, b0 = 0, B0 = 10,
marginal.likelihood = "Chib95")
log marginal likelihood = -220.6085
out2 :
call =
MCMCoprobitChange(formula = formula, m = 2, burnin = 100, mcmc = 100,
thin = 1, tune = c(0.5, 0.5, 0.5), verbose = 100, b0 = 0,
B0 = 10, marginal.likelihood = "Chib95")
log marginal likelihood = -241.987
>
> ## draw plots using the "right" model
> plotState(out1)
> plotChangepoint(out1)
>
>
>
>
>
> dev.off()
null device
1
>
|