Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
One-factorial ANOVA assessing spatial biasDescriptionThis function performs an one-factorial analysis of variance to test for spatial bias for a single array. The predictor variable is the average logged intensity of both channels and the response variable is the logged fold-change. Usageanovaspatial(obj,index,xN=5,yN=5,visu=FALSE) Arguments
DetailsThe function ValueThe return value is a list of summary statistics of the fitted model as produced by Author(s)Matthias E. Futschik (http://itb.biologie.hu-berlin.de/~futschik) See Also
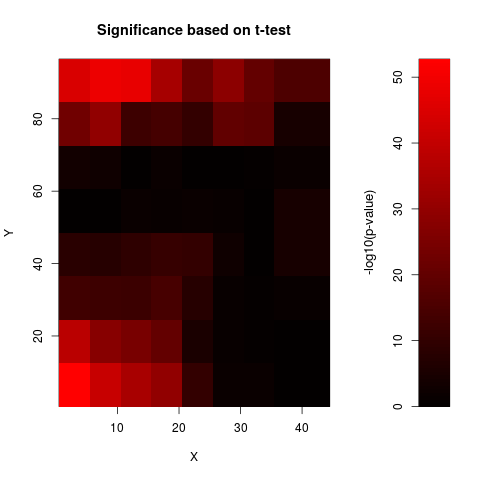
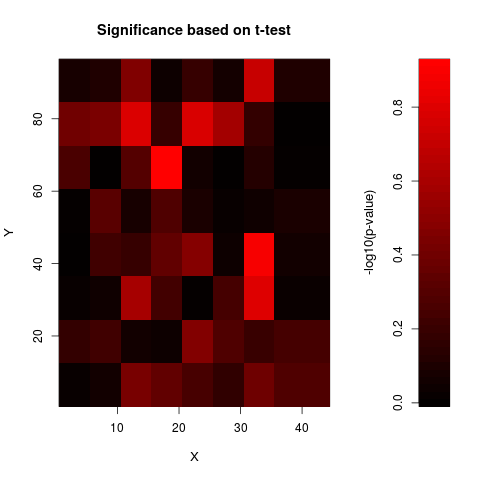
Examples# CHECK RAW DATA FOR SPATIAL BIAS data(sw) print(anovaspatial(sw,index=1,xN=8,yN=8,visu=TRUE)) # CHECK DATA NORMALISED BY OLIN FOR SPATIAL BIAS data(sw.olin) print(anovaspatial(sw.olin,index=1,xN=8,yN=8,visu=TRUE)) # note the different scale of the colour bar Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(OLIN)
Loading required package: locfit
locfit 1.5-9.1 2013-03-22
Loading required package: marray
Loading required package: limma
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/OLIN/anovaspatial.Rd_%03d_medium.png", width=480, height=480)
> ### Name: anovaspatial
> ### Title: One-factorial ANOVA assessing spatial bias
> ### Aliases: anovaspatial
> ### Keywords: models regression
>
> ### ** Examples
>
> # CHECK RAW DATA FOR SPATIAL BIAS
> data(sw)
> print(anovaspatial(sw,index=1,xN=8,yN=8,visu=TRUE))
Call:
lm(formula = as.vector(M) ~ factor(block) - 1)
Residuals:
Min 1Q Median 3Q Max
-3.04880 -0.29599 0.00474 0.28153 2.52965
Coefficients:
Estimate Std. Error t value Pr(>|t|)
factor(block)1 -0.9654777 0.0618191 -15.618 < 2e-16 ***
factor(block)2 -0.8163338 0.0618191 -13.205 < 2e-16 ***
factor(block)3 -0.4594468 0.0618191 -7.432 1.29e-13 ***
factor(block)4 -0.3616640 0.0618191 -5.850 5.28e-09 ***
factor(block)5 -0.0551087 0.0618191 -0.891 0.372739
factor(block)6 0.2269960 0.0618191 3.672 0.000244 ***
factor(block)7 0.6213434 0.0618191 10.051 < 2e-16 ***
factor(block)8 0.8845512 0.0618191 14.309 < 2e-16 ***
factor(block)9 -0.8452423 0.0618191 -13.673 < 2e-16 ***
factor(block)10 -0.6882680 0.0618191 -11.134 < 2e-16 ***
factor(block)11 -0.4471502 0.0618191 -7.233 5.59e-13 ***
factor(block)12 -0.3416448 0.0618191 -5.527 3.47e-08 ***
factor(block)13 0.0086121 0.0618191 0.139 0.889212
factor(block)14 0.1917838 0.0618191 3.102 0.001933 **
factor(block)15 0.7149640 0.0618191 11.565 < 2e-16 ***
factor(block)16 0.9256309 0.0618191 14.973 < 2e-16 ***
factor(block)17 -0.7742514 0.0618191 -12.524 < 2e-16 ***
factor(block)18 -0.6490230 0.0618191 -10.499 < 2e-16 ***
factor(block)19 -0.4425972 0.0618191 -7.160 9.52e-13 ***
factor(block)20 -0.3854886 0.0618191 -6.236 4.94e-10 ***
factor(block)21 -0.1530968 0.0618191 -2.477 0.013306 *
factor(block)22 0.0470847 0.0618191 0.762 0.446310
factor(block)23 0.4482664 0.0618191 7.251 4.90e-13 ***
factor(block)24 0.9157485 0.0618191 14.813 < 2e-16 ***
factor(block)25 -0.7126618 0.0618191 -11.528 < 2e-16 ***
factor(block)26 -0.5836128 0.0618191 -9.441 < 2e-16 ***
factor(block)27 -0.4903762 0.0618191 -7.932 2.74e-15 ***
factor(block)28 -0.4161683 0.0618191 -6.732 1.90e-11 ***
factor(block)29 -0.1285373 0.0618191 -2.079 0.037656 *
factor(block)30 0.1469066 0.0618191 2.376 0.017528 *
factor(block)31 0.4838255 0.0618191 7.826 6.32e-15 ***
factor(block)32 0.7691278 0.0618191 12.442 < 2e-16 ***
factor(block)33 -0.4054033 0.0618191 -6.558 6.13e-11 ***
factor(block)34 -0.2695021 0.0618191 -4.360 1.33e-05 ***
factor(block)35 -0.3482214 0.0618191 -5.633 1.89e-08 ***
factor(block)36 -0.4048784 0.0618191 -6.549 6.48e-11 ***
factor(block)37 -0.1490995 0.0618191 -2.412 0.015914 *
factor(block)38 0.0147999 0.0618191 0.239 0.810802
factor(block)39 0.4028761 0.0618191 6.517 8.03e-11 ***
factor(block)40 0.6024267 0.0618191 9.745 < 2e-16 ***
factor(block)41 -0.1635685 0.0618191 -2.646 0.008178 **
factor(block)42 -0.1188741 0.0618191 -1.923 0.054557 .
factor(block)43 -0.1313807 0.0618191 -2.125 0.033625 *
factor(block)44 -0.1901258 0.0618191 -3.076 0.002115 **
factor(block)45 -0.1154434 0.0618191 -1.867 0.061911 .
factor(block)46 -0.0167989 0.0618191 -0.272 0.785833
factor(block)47 0.5785432 0.0618191 9.359 < 2e-16 ***
factor(block)48 0.7047983 0.0618191 11.401 < 2e-16 ***
factor(block)49 0.1376708 0.0618191 2.227 0.026001 *
factor(block)50 0.1014679 0.0618191 1.641 0.100797
factor(block)51 -0.0692517 0.0618191 -1.120 0.262680
factor(block)52 0.0362919 0.0618191 0.587 0.557192
factor(block)53 0.0353528 0.0618191 0.572 0.567437
factor(block)54 0.0971955 0.0618191 1.572 0.115967
factor(block)55 0.5620149 0.0618191 9.091 < 2e-16 ***
factor(block)56 0.5884679 0.0618191 9.519 < 2e-16 ***
factor(block)57 -0.0008855 0.0460773 -0.019 0.984668
factor(block)58 -0.0136021 0.0460773 -0.295 0.767854
factor(block)59 -0.0955905 0.0460773 -2.075 0.038088 *
factor(block)60 -0.1961225 0.0460773 -4.256 2.12e-05 ***
factor(block)61 -0.1929842 0.0460773 -4.188 2.87e-05 ***
factor(block)62 -0.1146842 0.0460773 -2.489 0.012851 *
factor(block)63 0.1908167 0.0460773 4.141 3.52e-05 ***
factor(block)64 0.3764879 0.0460773 8.171 4.02e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.4788 on 4160 degrees of freedom
Multiple R-squared: 0.4655, Adjusted R-squared: 0.4573
F-statistic: 56.6 on 64 and 4160 DF, p-value: < 2.2e-16
>
>
> # CHECK DATA NORMALISED BY OLIN FOR SPATIAL BIAS
> data(sw.olin)
> print(anovaspatial(sw.olin,index=1,xN=8,yN=8,visu=TRUE))
Call:
lm(formula = as.vector(M) ~ factor(block) - 1)
Residuals:
Min 1Q Median 3Q Max
-3.2153 -0.1665 0.0138 0.1789 2.4621
Coefficients:
Estimate Std. Error t value Pr(>|t|)
factor(block)1 -0.0033065 0.0498601 -0.066 0.947
factor(block)2 -0.0215496 0.0498601 -0.432 0.666
factor(block)3 0.0033983 0.0498601 0.068 0.946
factor(block)4 -0.0007232 0.0498601 -0.015 0.988
factor(block)5 0.0013500 0.0498601 0.027 0.978
factor(block)6 0.0300595 0.0498601 0.603 0.547
factor(block)7 -0.0419285 0.0498601 -0.841 0.400
factor(block)8 0.0106547 0.0498601 0.214 0.831
factor(block)9 0.0086288 0.0498601 0.173 0.863
factor(block)10 0.0263100 0.0498601 0.528 0.598
factor(block)11 0.0069643 0.0498601 0.140 0.889
factor(block)12 0.0265694 0.0498601 0.533 0.594
factor(block)13 0.0348534 0.0498601 0.699 0.485
factor(block)14 -0.0005567 0.0498601 -0.011 0.991
factor(block)15 0.0447804 0.0498601 0.898 0.369
factor(block)16 0.0134916 0.0498601 0.271 0.787
factor(block)17 -0.0446339 0.0498601 -0.895 0.371
factor(block)18 -0.0082794 0.0498601 -0.166 0.868
factor(block)19 0.0569828 0.0498601 1.143 0.253
factor(block)20 -0.0227180 0.0498601 -0.456 0.649
factor(block)21 -0.0106875 0.0498601 -0.214 0.830
factor(block)22 -0.0336463 0.0498601 -0.675 0.500
factor(block)23 -0.0692836 0.0498601 -1.390 0.165
factor(block)24 0.0462709 0.0498601 0.928 0.353
factor(block)25 -0.0375197 0.0498601 -0.752 0.452
factor(block)26 0.0059531 0.0498601 0.119 0.905
factor(block)27 -0.0272394 0.0498601 -0.546 0.585
factor(block)28 -0.0377998 0.0498601 -0.758 0.448
factor(block)29 0.0314399 0.0498601 0.631 0.528
factor(block)30 0.0774899 0.0498601 1.554 0.120
factor(block)31 -0.0224376 0.0498601 -0.450 0.653
factor(block)32 -0.0062391 0.0498601 -0.125 0.900
factor(block)33 0.0293269 0.0498601 0.588 0.556
factor(block)34 0.0473647 0.0498601 0.950 0.342
factor(block)35 -0.0016106 0.0498601 -0.032 0.974
factor(block)36 -0.0480839 0.0498601 -0.964 0.335
factor(block)37 -0.0118535 0.0498601 -0.238 0.812
factor(block)38 -0.0078246 0.0498601 -0.157 0.875
factor(block)39 -0.0686859 0.0498601 -1.378 0.168
factor(block)40 -0.0223675 0.0498601 -0.449 0.654
factor(block)41 -0.0206801 0.0498601 -0.415 0.678
factor(block)42 -0.0319311 0.0498601 -0.640 0.522
factor(block)43 0.0286646 0.0498601 0.575 0.565
factor(block)44 -0.0060567 0.0498601 -0.121 0.903
factor(block)45 0.0032303 0.0498601 0.065 0.948
factor(block)46 0.0006503 0.0498601 0.013 0.990
factor(block)47 0.0563123 0.0498601 1.129 0.259
factor(block)48 0.0095686 0.0498601 0.192 0.848
factor(block)49 0.0410204 0.0498601 0.823 0.411
factor(block)50 0.0238266 0.0498601 0.478 0.633
factor(block)51 -0.0702298 0.0498601 -1.409 0.159
factor(block)52 0.0754324 0.0498601 1.513 0.130
factor(block)53 0.0070427 0.0498601 0.141 0.888
factor(block)54 0.0155062 0.0498601 0.311 0.756
factor(block)55 0.0218938 0.0498601 0.439 0.661
factor(block)56 -0.0651155 0.0498601 -1.306 0.192
factor(block)57 -0.0237302 0.0371635 -0.639 0.523
factor(block)58 0.0208438 0.0371635 0.561 0.575
factor(block)59 0.0036944 0.0371635 0.099 0.921
factor(block)60 -0.0056216 0.0371635 -0.151 0.880
factor(block)61 -0.0084312 0.0371635 -0.227 0.821
factor(block)62 0.0011146 0.0371635 0.030 0.976
factor(block)63 0.0005883 0.0371635 0.016 0.987
factor(block)64 -0.0099045 0.0371635 -0.267 0.790
Residual standard error: 0.3862 on 4160 degrees of freedom
Multiple R-squared: 0.006707, Adjusted R-squared: -0.008574
F-statistic: 0.4389 on 64 and 4160 DF, p-value: 1
> # note the different scale of the colour bar
>
>
>
>
>
>
> dev.off()
null device
1
>
|