Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Local intensity-dependent normalisation of two-colour microarraysDescriptionThis functions performs local intensity-dependent normalisation (LIN) Usagelin(object,X=NA,Y=NA,alpha=0.3,iter=2,scale=TRUE,weights=NA,bg.corr="subtract",...) Arguments
DetailsLIN is based on the same normalisation scheme as OLIN, but does not incorporate
optimisation of model parameters. The function The smoothing parameter If the normalisation should be based on set of genes assumed to be not differentially expressed (house-keeping genes), weights can be used for local regression. In this case, all weights should be set to zero except for the house-keeping genes for which weights are set to one. In order to achieve a reliable regression, it is important, however, that there is a sufficient number of house-keeping genes that cover the whole expression range and are spotted accross the whole array. ValueObject of class “marrayNorm” with normalised logged ratios Author(s)Matthias E. Futschik (http://itb.biologie.hu-berlin.de/~futschik) References
See Also
Examples
# LOADING DATA
data(sw)
data(sw.xy)
# LOCAL INTENSITY-DEPENDENT NORMALISATION
norm.lin <- lin(sw,X=sw.xy$X,Y=sw.xy$Y)
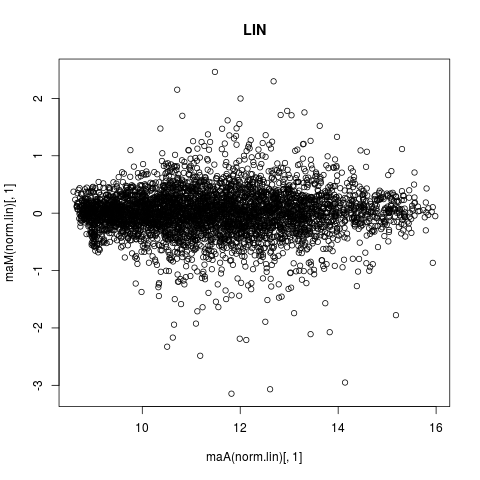
# MA-PLOT OF NORMALISATION RESULTS OF FIRST ARRAY
plot(maA(norm.lin)[,1],maM(norm.lin)[,1],main="LIN")
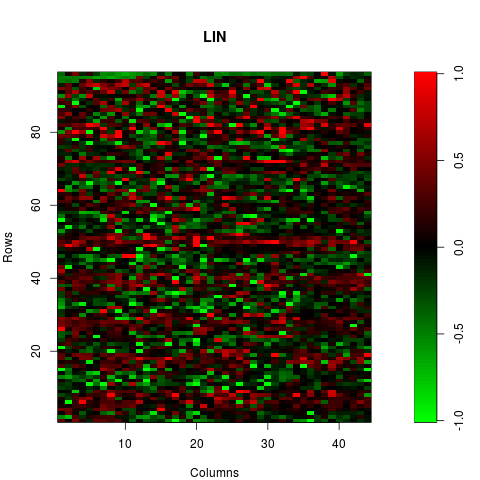
# CORRESPONDING MXY-PLOT
mxy.plot(maM(norm.lin)[,1],Ngc=maNgc(norm.lin),Ngr=maNgr(norm.lin),
Nsc=maNsc(norm.lin),Nsr=maNsr(norm.lin),main="LIN")
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(OLIN)
Loading required package: locfit
locfit 1.5-9.1 2013-03-22
Loading required package: marray
Loading required package: limma
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/OLIN/lin.Rd_%03d_medium.png", width=480, height=480)
> ### Name: lin
> ### Title: Local intensity-dependent normalisation of two-colour
> ### microarrays
> ### Aliases: lin
> ### Keywords: utilities regression
>
> ### ** Examples
>
>
>
>
> # LOADING DATA
> data(sw)
> data(sw.xy)
>
> # LOCAL INTENSITY-DEPENDENT NORMALISATION
> norm.lin <- lin(sw,X=sw.xy$X,Y=sw.xy$Y)
>
> # MA-PLOT OF NORMALISATION RESULTS OF FIRST ARRAY
> plot(maA(norm.lin)[,1],maM(norm.lin)[,1],main="LIN")
>
> # CORRESPONDING MXY-PLOT
> mxy.plot(maM(norm.lin)[,1],Ngc=maNgc(norm.lin),Ngr=maNgr(norm.lin),
+ Nsc=maNsc(norm.lin),Nsr=maNsr(norm.lin),main="LIN")
>
>
>
>
>
>
>
> dev.off()
null device
1
>
|