Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Compare balance graphically of a continuous covariate as part of a PSADescriptionGiven predefined strata and two level treatment for a continuous covariate from a propensity score analysis,
Usage
box.psa(continuous, treatment = NULL, strata = NULL, boxwex = 0.17,
offset = 0.17,
col = c("yellow", "orange", "black", "red", "darkorange3"),
xlab = "Stratum", legend.xy = NULL,
legend.labels = NULL, pts = TRUE,
balance = FALSE, trim = 0, B = 1000, ...)
Arguments
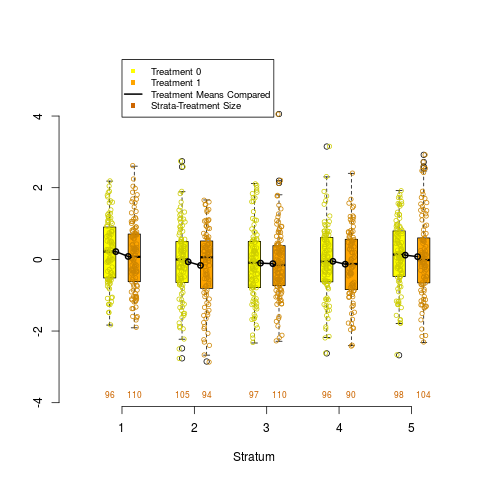
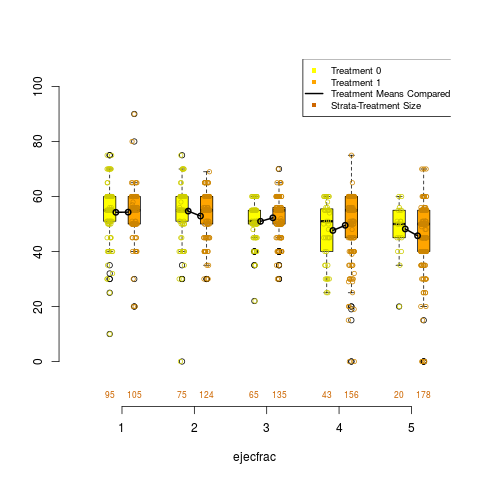
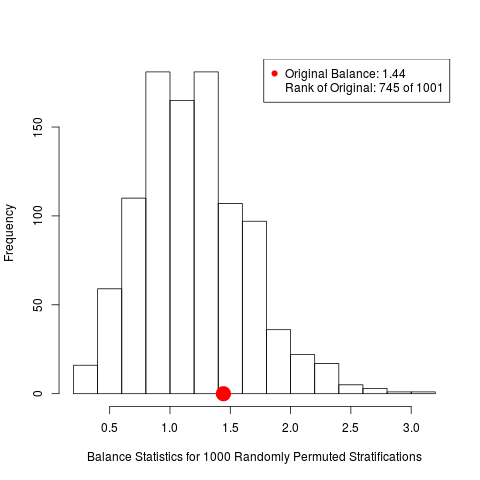
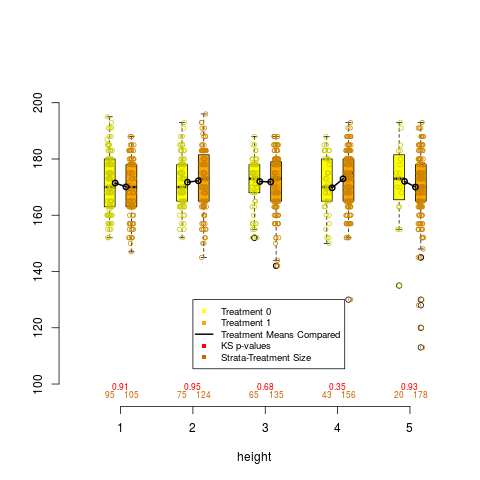
DetailsDraws a pair of side by side boxplots for each stratum of a propensity score analysis. This allows visual comparisons within strata of the distribution of the given continuous covariate, and comparisons between strata as well. The number of observations in each boxplot are given below each box, and the means of paired treatment and control groups are connected. Author(s)James E. Helmreich James.Helmreich@Marist.edu Robert M. Pruzek RMPruzek@yahoo.com See Also
Examples
continuous<-rnorm(1000)
treatment<-sample(c(0,1),1000,replace=TRUE)
strata<-sample(5,1000,replace=TRUE)
box.psa(continuous, treatment, strata)
data(lindner)
attach(lindner)
lindner.ps <- glm(abcix ~ stent + height + female +
diabetic + acutemi + ejecfrac + ves1proc,
data = lindner, family = binomial)
ps<-lindner.ps$fitted
lindner.s5 <- as.numeric(cut(ps, quantile(ps, seq(0, 1, 1/5)),
include.lowest = TRUE, labels = FALSE))
box.psa(ejecfrac, abcix, lindner.s5, xlab = "ejecfrac",
legend.xy = c(3.5,110))
lindner.s10 <- as.numeric(cut(ps, quantile(ps, seq(0, 1, 1/5)),
include.lowest = TRUE, labels = FALSE))
box.psa(height, abcix, lindner.s10, xlab="height",
boxwex = .15, offset = .15, legend.xy = c(2,130), balance = TRUE)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(PSAgraphics)
Loading required package: rpart
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/PSAgraphics/box.psa.Rd_%03d_medium.png", width=480, height=480)
> ### Name: box.psa
> ### Title: Compare balance graphically of a continuous covariate as part of
> ### a PSA
> ### Aliases: box.psa
> ### Keywords: hplot htest
>
> ### ** Examples
>
> continuous<-rnorm(1000)
> treatment<-sample(c(0,1),1000,replace=TRUE)
> strata<-sample(5,1000,replace=TRUE)
> box.psa(continuous, treatment, strata)
Warning messages:
1: In xy.coords(x, y) : NAs introduced by coercion
2: In xy.coords(x, y) : NAs introduced by coercion
>
> data(lindner)
> attach(lindner)
> lindner.ps <- glm(abcix ~ stent + height + female +
+ diabetic + acutemi + ejecfrac + ves1proc,
+ data = lindner, family = binomial)
> ps<-lindner.ps$fitted
> lindner.s5 <- as.numeric(cut(ps, quantile(ps, seq(0, 1, 1/5)),
+ include.lowest = TRUE, labels = FALSE))
> box.psa(ejecfrac, abcix, lindner.s5, xlab = "ejecfrac",
+ legend.xy = c(3.5,110))
Warning messages:
1: In xy.coords(x, y) : NAs introduced by coercion
2: In xy.coords(x, y) : NAs introduced by coercion
>
> lindner.s10 <- as.numeric(cut(ps, quantile(ps, seq(0, 1, 1/5)),
+ include.lowest = TRUE, labels = FALSE))
> box.psa(height, abcix, lindner.s10, xlab="height",
+ boxwex = .15, offset = .15, legend.xy = c(2,130), balance = TRUE)
Press <enter> for bar chart...
Warning messages:
1: In ks.test(continuous[treatment == unique(treatment)[1] & strat.f == :
cannot compute exact p-value with ties
2: In ks.test(continuous[treatment == unique(treatment)[1] & strat.f == :
cannot compute exact p-value with ties
3: In ks.test(continuous[treatment == unique(treatment)[1] & strat.f == :
cannot compute exact p-value with ties
4: In ks.test(continuous[treatment == unique(treatment)[1] & strat.f == :
cannot compute exact p-value with ties
5: In ks.test(continuous[treatment == unique(treatment)[1] & strat.f == :
cannot compute exact p-value with ties
6: In xy.coords(x, y) : NAs introduced by coercion
7: In xy.coords(x, y) : NAs introduced by coercion
>
>
>
>
>
> dev.off()
null device
1
>
|