Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Expectation conditional maximization of negative binomial HMM parameters using forward-backward algorithmDescriptionGiven an input read count vector of integers, the function optimizes the parameters for the negative binomial HMM of K hidden states using expectation conditional maximization with forward-backward algorithm to acheive the exact inference. Usagenbh_em(count, TRANS, alpha, beta, NBH_NIT_MAX = 250, NBH_TOL = 1e-05, MAXALPHA = 1e+07, MAXBETA = 1e+07) Arguments
DetailsGiven a K-state HMM with NB emission (NBH), the goal is to maximize the likelihood function with respect to the parameters comprising of α_k and β_k for the K NB components and the transition probabilities A_jk between any state j and k, which are the priors p(z=k). Because there is no analytical solution for the maximum likelihood (ML) estimators of the above quantities, a modified EM procedures called Expectation Conditional Maximization is employed (Meng and Rubin, 1994). In E-step, the posterior probability is evaluated by forward-backward algorithm using NB density functions with initialized alpha, beta, and TRANS. In the CM step, A_jk is evaluated first followed by Newton updates of α_k and β_k. EM iteration terminates when the percetnage of increase of log likelihood drop below ValueA list containing:
Author(s)Yue Li ReferencesRabiner, L. R. (1989). A tutorial on hidden Markov models and selected applications in speech recognition (Vol. 77, pp. 257-286). Presented at the Proceedings of the IEEE. doi:10.1109/5.18626 Christopher Bishop. Pattern recognition and machine learning. Number 605-631 in Information Science and Statisitcs. Springer Science, 2006. X. L. Meng, D. B. Rubin, Maximum likelihood estimation via the ECM algorithm: A general framework, Biometrika, 80(2):267-278 (1993). J. A. Fessler, A. O. Hero, Space-alternating generalized expectation-maximization algorithm, IEEE Tr. on Signal Processing, 42(10):2664 -2677 (1994). Capp'e, O. (2001). H2M : A set of MATLAB/OCTAVE functions for the EM estimation of mixtures and hidden Markov models. (http://perso.telecom-paristech.fr/cappe/h2m/) See Also
Examples
# Simulate data
TRANS_s <- matrix(c(0.9, 0.1, 0.3, 0.7), nrow=2, byrow=TRUE)
alpha_s <- c(2, 4)
beta_s <- c(1, 0.25)
Total <- 100
x <- nbh_gen(TRANS_s, alpha_s, beta_s, Total);
count <- x$count
label <- x$label
Total <- length(count)
# dummy initialization
TRANS0 <- matrix(rep(0.5,4), 2)
alpha0 <- c(1, 20)
beta0 <- c(1, 1)
NIT_MAX <- 50
TOL <- 1e-100
nbh <- nbh_em(count, TRANS0, alpha0, beta0, NIT_MAX, TOL)
map.accuracy <- length(which(max.col(nbh$postprob) == label))/Total
vit <- nbh_vit(count, nbh$TRANS, nbh$alpha, nbh$beta)
vit.accuracy <- length(which(vit$class == label))/Total
# Plots
par(mfrow=c(2,2), cex.lab=1.2, cex.main=1.2)
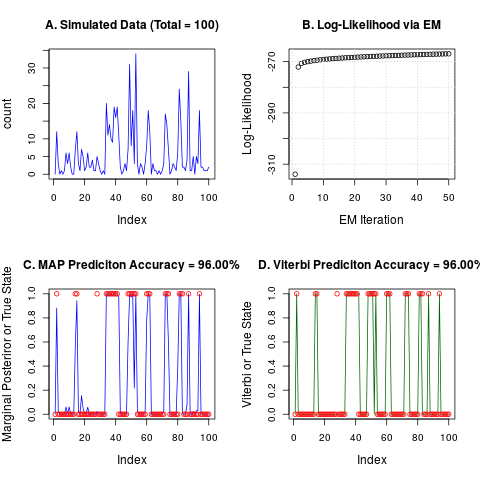
plot(count, col="blue", type="l", main=sprintf("A. Simulated Data (Total = %i)",Total))
plot(as.numeric(nbh$logl), xlab="EM Iteration", ylab="Log-Likelihood",
main="B. Log-Likelihood via EM");grid()
# Marginal postprob
plot(nbh$postprob[,2], col="blue", type="l", ylim = c(0,1),
ylab="Marginal Posteriror or True State")
points(label-1, col="red")
title(main = sprintf("C. MAP Prediciton Accuracy = %.2f%s", 100 * map.accuracy, "%"))
# Viterbi states
plot(vit$class - 1, col="dark green", type="l", ylim = c(0,1),
ylab="Viterbi or True State")
points(label-1, col="red")
title(main = sprintf("D. Viterbi Prediciton Accuracy = %.2f%s", 100 * vit.accuracy, "%"))
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(RIPSeeker)
Loading required package: S4Vectors
Loading required package: stats4
Loading required package: BiocGenerics
Loading required package: parallel
Attaching package: 'BiocGenerics'
The following objects are masked from 'package:parallel':
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLB
The following objects are masked from 'package:stats':
IQR, mad, xtabs
The following objects are masked from 'package:base':
Filter, Find, Map, Position, Reduce, anyDuplicated, append,
as.data.frame, cbind, colnames, do.call, duplicated, eval, evalq,
get, grep, grepl, intersect, is.unsorted, lapply, lengths, mapply,
match, mget, order, paste, pmax, pmax.int, pmin, pmin.int, rank,
rbind, rownames, sapply, setdiff, sort, table, tapply, union,
unique, unsplit
Attaching package: 'S4Vectors'
The following objects are masked from 'package:base':
colMeans, colSums, expand.grid, rowMeans, rowSums
Loading required package: IRanges
Loading required package: GenomicRanges
Loading required package: GenomeInfoDb
Loading required package: SummarizedExperiment
Loading required package: Biobase
Welcome to Bioconductor
Vignettes contain introductory material; view with
'browseVignettes()'. To cite Bioconductor, see
'citation("Biobase")', and for packages 'citation("pkgname")'.
Loading required package: Rsamtools
Loading required package: Biostrings
Loading required package: XVector
Loading required package: GenomicAlignments
Loading required package: rtracklayer
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/RIPSeeker/nbh_em.Rd_%03d_medium.png", width=480, height=480)
> ### Name: nbh_em
> ### Title: Expectation conditional maximization of negative binomial HMM
> ### parameters using forward-backward algorithm
> ### Aliases: nbh_em
>
> ### ** Examples
>
> # Simulate data
> TRANS_s <- matrix(c(0.9, 0.1, 0.3, 0.7), nrow=2, byrow=TRUE)
> alpha_s <- c(2, 4)
> beta_s <- c(1, 0.25)
> Total <- 100
>
> x <- nbh_gen(TRANS_s, alpha_s, beta_s, Total);
>
> count <- x$count
> label <- x$label
>
> Total <- length(count)
>
> # dummy initialization
> TRANS0 <- matrix(rep(0.5,4), 2)
>
> alpha0 <- c(1, 20)
>
> beta0 <- c(1, 1)
>
> NIT_MAX <- 50
> TOL <- 1e-100
> nbh <- nbh_em(count, TRANS0, alpha0, beta0, NIT_MAX, TOL)
Iteration 0: -313.917
Iteration 1: -272.103
Iteration 2: -270.678
Iteration 3: -270.271
Iteration 4: -270.008
Iteration 5: -269.789
Iteration 6: -269.600
Iteration 7: -269.434
Iteration 8: -269.289
Iteration 9: -269.159
Iteration 10: -269.041
Iteration 11: -268.934
Iteration 12: -268.836
Iteration 13: -268.745
Iteration 14: -268.659
Iteration 15: -268.579
Iteration 16: -268.503
Iteration 17: -268.430
Iteration 18: -268.360
Iteration 19: -268.294
Iteration 20: -268.229
Iteration 21: -268.167
Iteration 22: -268.107
Iteration 23: -268.048
Iteration 24: -267.991
Iteration 25: -267.936
Iteration 26: -267.882
Iteration 27: -267.829
Iteration 28: -267.778
Iteration 29: -267.728
Iteration 30: -267.678
Iteration 31: -267.631
Iteration 32: -267.584
Iteration 33: -267.538
Iteration 34: -267.493
Iteration 35: -267.450
Iteration 36: -267.407
Iteration 37: -267.366
Iteration 38: -267.325
Iteration 39: -267.286
Iteration 40: -267.247
Iteration 41: -267.210
Iteration 42: -267.173
Iteration 43: -267.138
Iteration 44: -267.103
Iteration 45: -267.070
Iteration 46: -267.037
Iteration 47: -267.005
Iteration 48: -266.975
Iteration 49: -266.945
>
> map.accuracy <- length(which(max.col(nbh$postprob) == label))/Total
>
> vit <- nbh_vit(count, nbh$TRANS, nbh$alpha, nbh$beta)
>
> vit.accuracy <- length(which(vit$class == label))/Total
>
> # Plots
> par(mfrow=c(2,2), cex.lab=1.2, cex.main=1.2)
>
> plot(count, col="blue", type="l", main=sprintf("A. Simulated Data (Total = %i)",Total))
>
> plot(as.numeric(nbh$logl), xlab="EM Iteration", ylab="Log-Likelihood",
+ main="B. Log-Likelihood via EM");grid()
>
>
> # Marginal postprob
> plot(nbh$postprob[,2], col="blue", type="l", ylim = c(0,1),
+ ylab="Marginal Posteriror or True State")
> points(label-1, col="red")
> title(main = sprintf("C. MAP Prediciton Accuracy = %.2f%s", 100 * map.accuracy, "%"))
>
>
> # Viterbi states
> plot(vit$class - 1, col="dark green", type="l", ylim = c(0,1),
+ ylab="Viterbi or True State")
> points(label-1, col="red")
> title(main = sprintf("D. Viterbi Prediciton Accuracy = %.2f%s", 100 * vit.accuracy, "%"))
>
>
>
>
>
>
>
> dev.off()
null device
1
>
|