Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Plot SK and SK.nest ObjectsDescriptionS3 method to plot Usage
## S3 method for class 'SK'
plot(x,
pch=19,
col=NULL,
xlab=NULL,
ylab=NULL,
xlim=NULL,
ylim=NULL,
id.lab=NULL,
id.las=1,
id.col=TRUE,
rl=TRUE,
rl.lty=3,
rl.col='gray',
mm=TRUE,
mm.lty=1,
title="Means grouped by color(s)", ...)
Arguments
DetailsThe Author(s)Enio Jelihovschi (eniojelihovs@gmail.com) ReferencesMurrell, P. 2005. R Graphics. Chapman & Hall/CRC Press. See Also
Examples
##
## Examples: Completely Randomized Design (CRD)
## More details: demo(package='ScottKnott')
##
library(ScottKnott)
data(CRD2)
## From: vectors x and y
sk1 <- with(CRD2,
SK(x=x,
y=y,
model='y ~ x',
which='x'))
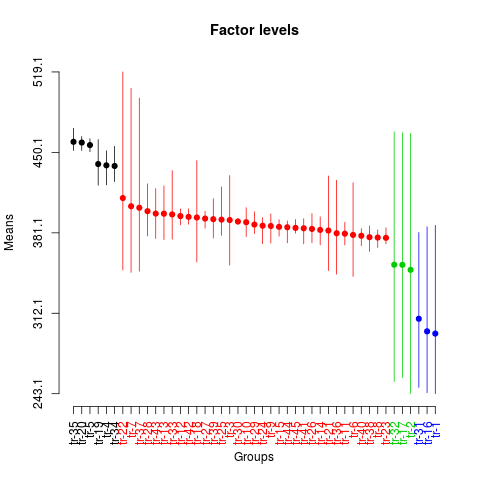
plot(sk1,
id.las=2,
rl=FALSE,
title='Factor levels')
## From: design matrix (dm) and response variable (y)
sk2 <- with(CRD2,
SK(x=dm,
y=y,
model='y ~ x',
which='x'))
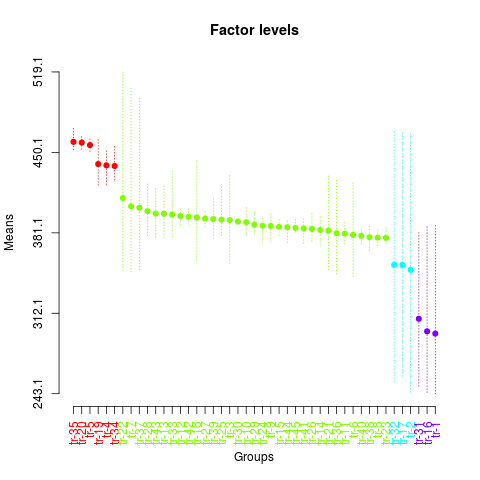
plot(sk2,
col=rainbow(max(sk2$groups)),
mm.lty=3,
id.las=2,
rl=FALSE,
title='Factor levels')
## From: data.frame (dfm)
sk3 <- with(CRD2,
SK(x=dfm,
model='y ~ x',
which='x'))
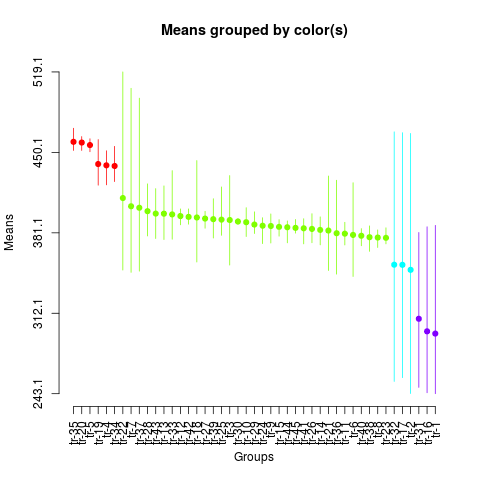
plot(sk3,
col=rainbow(max(sk3$groups)),
id.las=2,
id.col=FALSE,
rl=FALSE)
## From: aov
av <- with(CRD2,
aov(y ~ x,
data=dfm))
summary(av)
sk4 <- with(CRD2,
SK(x=av,
which='x'))
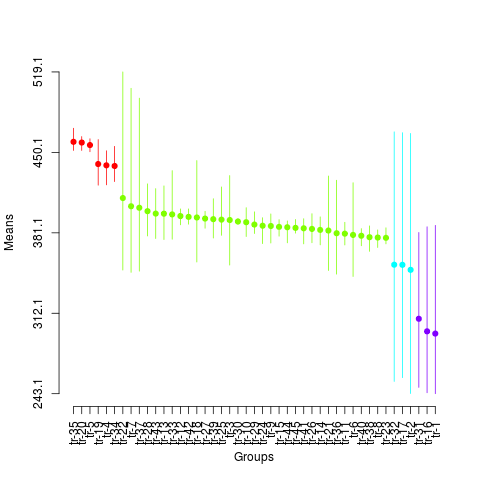
plot(sk4,
col=rainbow(max(sk4$groups)),
rl=FALSE,
id.las=2,
id.col=FALSE,
title=NULL)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(ScottKnott)
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/ScottKnott/plot.SK.Rd_%03d_medium.png", width=480, height=480)
> ### Name: plot.SK
> ### Title: Plot SK and SK.nest Objects
> ### Aliases: plot.SK
> ### Keywords: package htest univar tree design
>
> ### ** Examples
>
> ##
> ## Examples: Completely Randomized Design (CRD)
> ## More details: demo(package='ScottKnott')
> ##
>
> library(ScottKnott)
> data(CRD2)
>
> ## From: vectors x and y
> sk1 <- with(CRD2,
+ SK(x=x,
+ y=y,
+ model='y ~ x',
+ which='x'))
>
> plot(sk1,
+ id.las=2,
+ rl=FALSE,
+ title='Factor levels')
>
> ## From: design matrix (dm) and response variable (y)
> sk2 <- with(CRD2,
+ SK(x=dm,
+ y=y,
+ model='y ~ x',
+ which='x'))
> plot(sk2,
+ col=rainbow(max(sk2$groups)),
+ mm.lty=3,
+ id.las=2,
+ rl=FALSE,
+ title='Factor levels')
>
> ## From: data.frame (dfm)
> sk3 <- with(CRD2,
+ SK(x=dfm,
+ model='y ~ x',
+ which='x'))
>
> plot(sk3,
+ col=rainbow(max(sk3$groups)),
+ id.las=2,
+ id.col=FALSE,
+ rl=FALSE)
>
> ## From: aov
> av <- with(CRD2,
+ aov(y ~ x,
+ data=dfm))
> summary(av)
Df Sum Sq Mean Sq F value Pr(>F)
x 44 209136 4753 3.273 7.69e-08 ***
Residuals 135 196045 1452
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
>
> sk4 <- with(CRD2,
+ SK(x=av,
+ which='x'))
>
> plot(sk4,
+ col=rainbow(max(sk4$groups)),
+ rl=FALSE,
+ id.las=2,
+ id.col=FALSE,
+ title=NULL)
>
>
>
>
>
> dev.off()
null device
1
>
|