Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Fitting Contamination ModelDescriptionProvides ML estimates of a Gaussian contamination model. Usage
ml.est (y, x=NULL, model = "LN", lambda=3, w=0.05,
lambda.fix=FALSE, w.fix=FALSE, eps=1e-7,
max.iter=500, t.outl=0.5, graph=FALSE)
Arguments
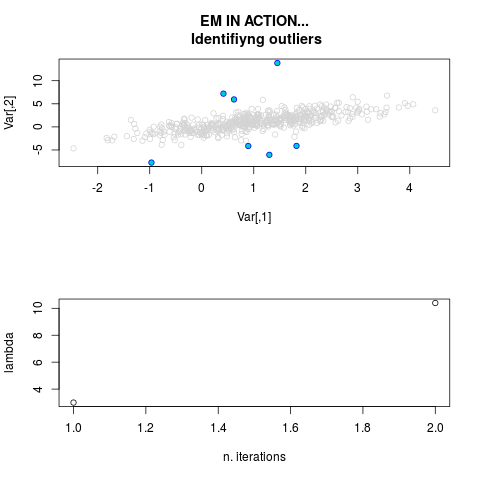
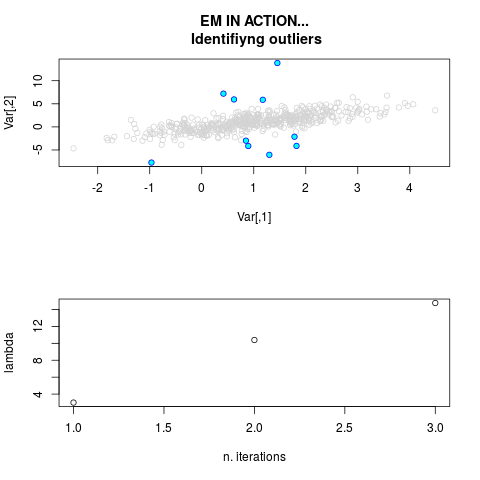
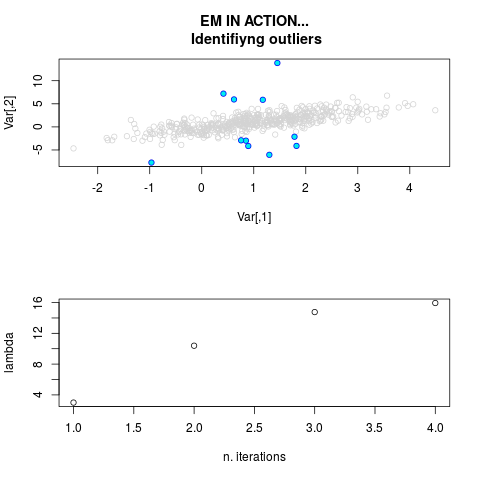
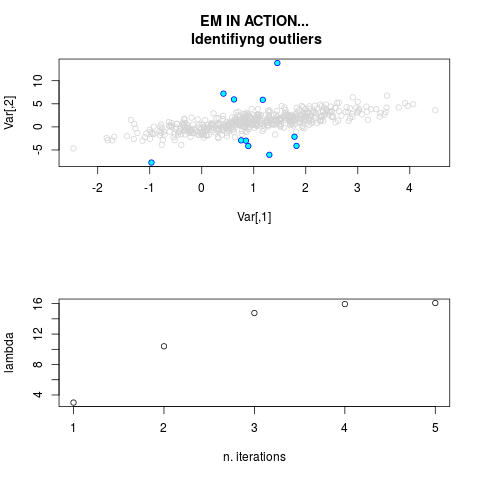
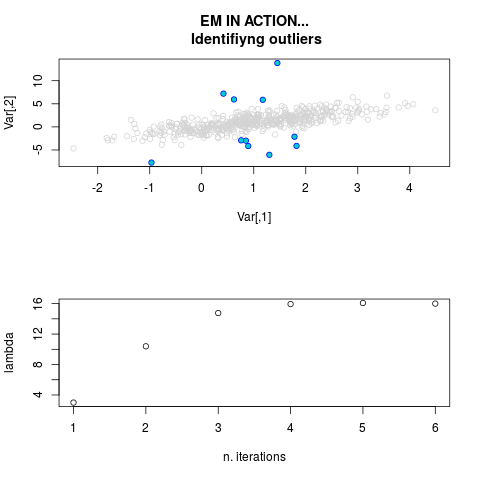
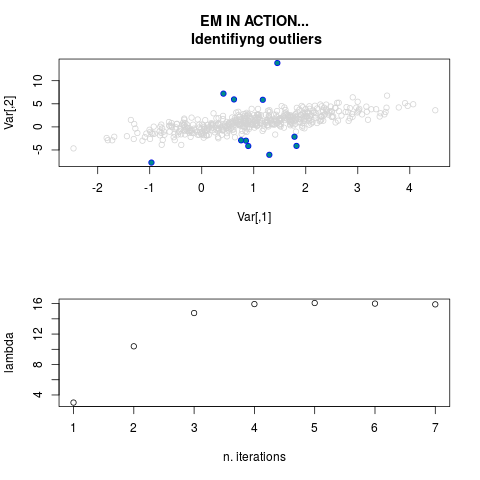
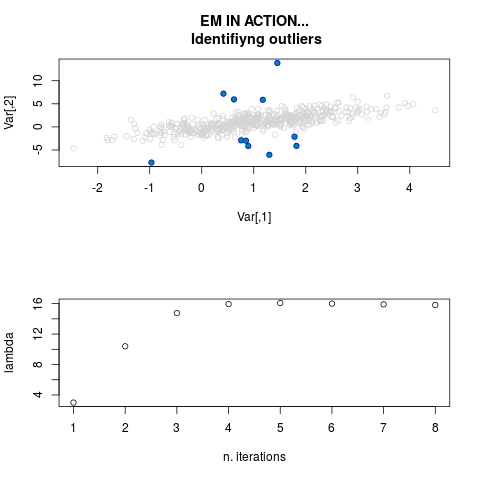
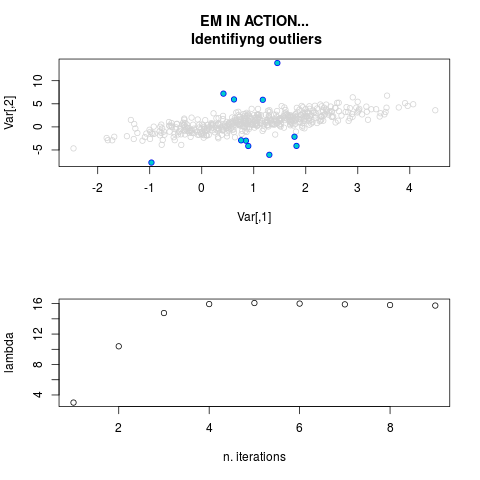
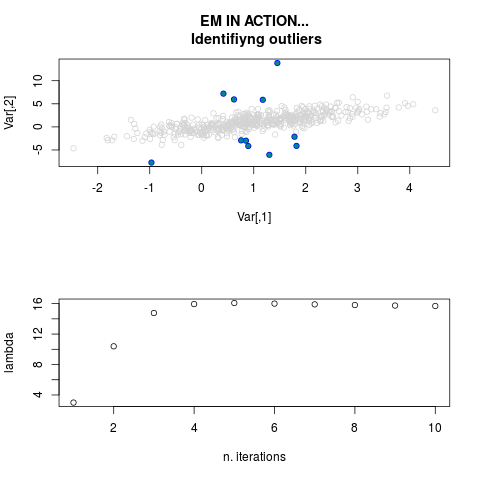
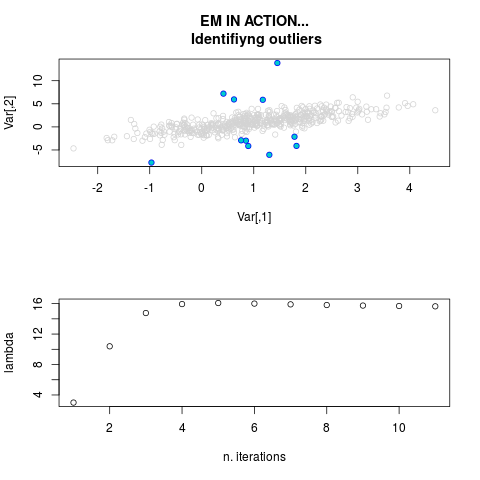
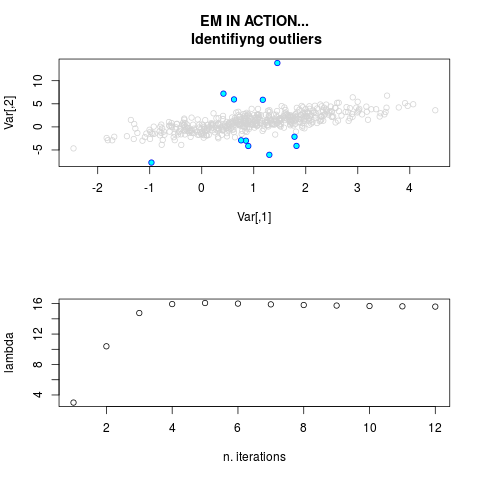
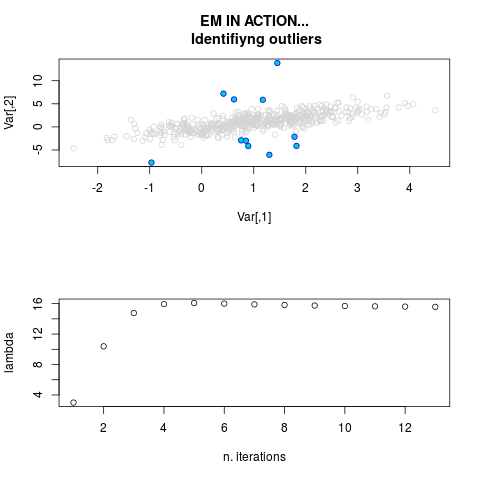
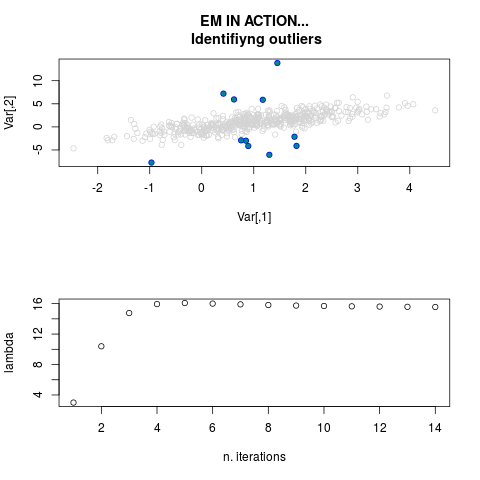
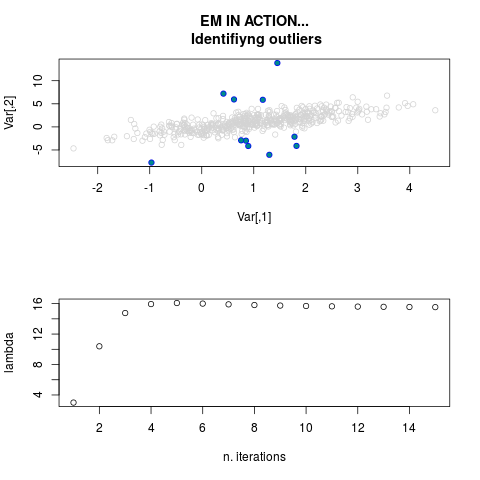
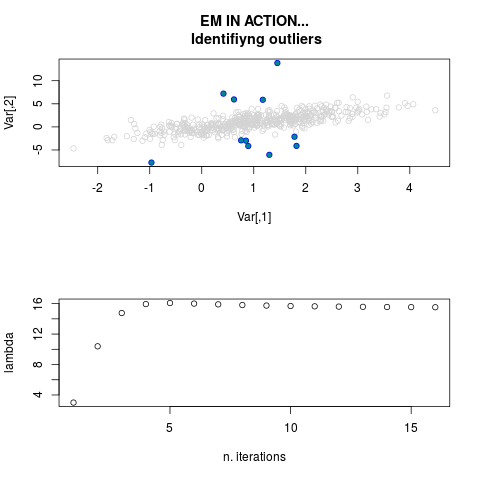
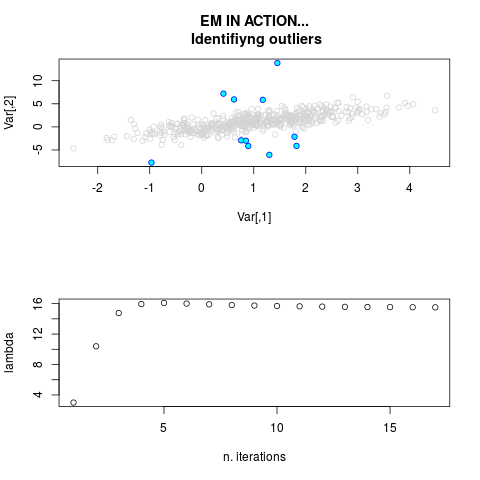
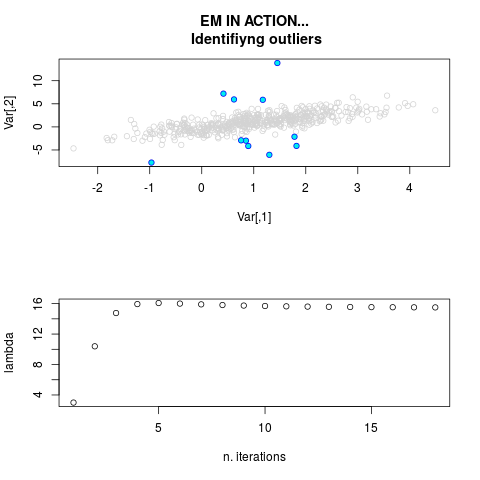
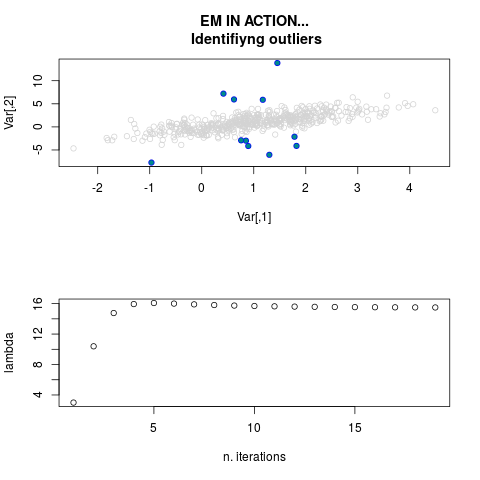
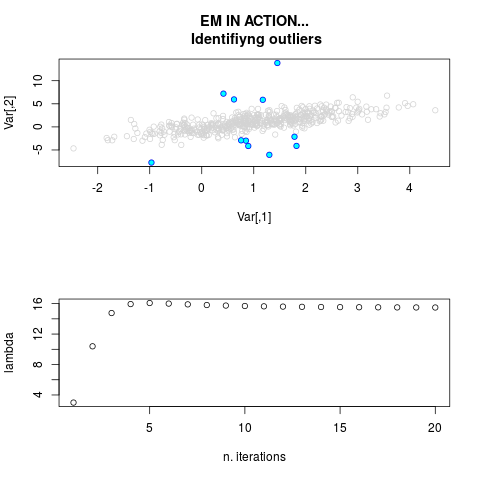
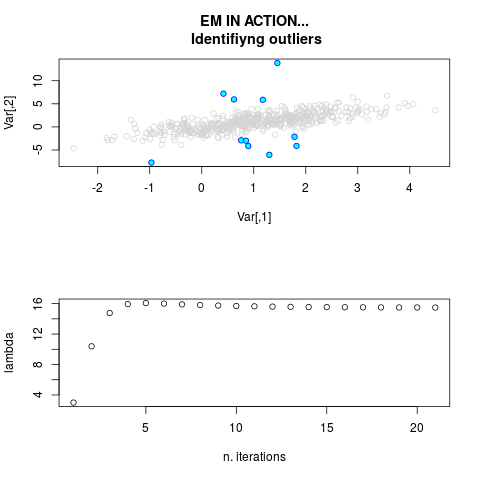
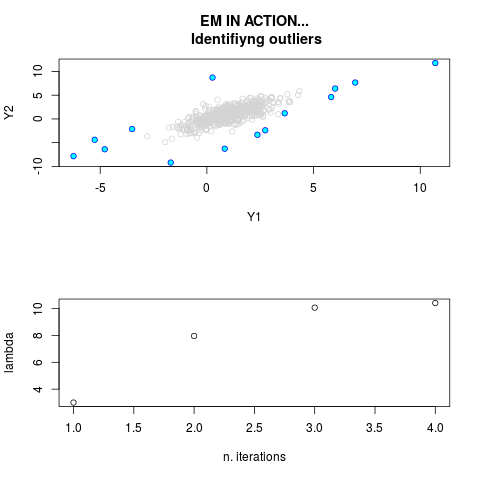
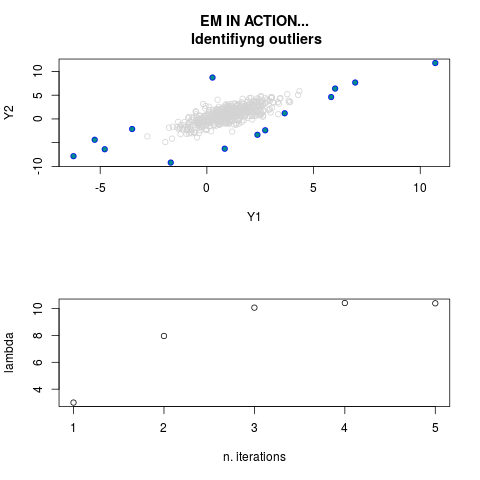
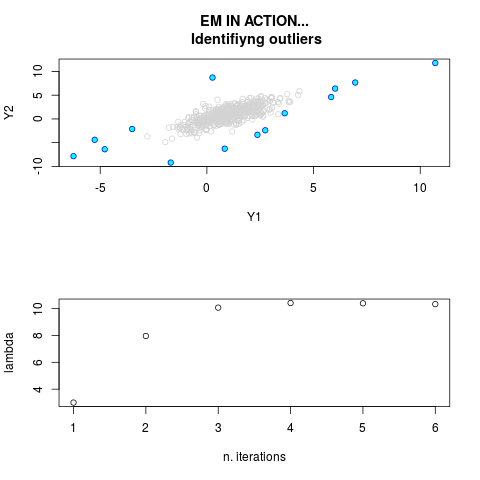
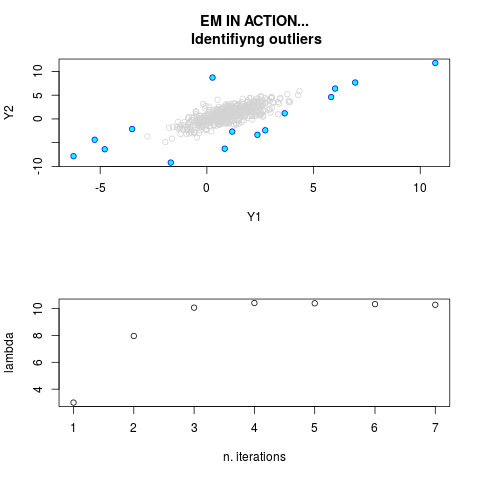
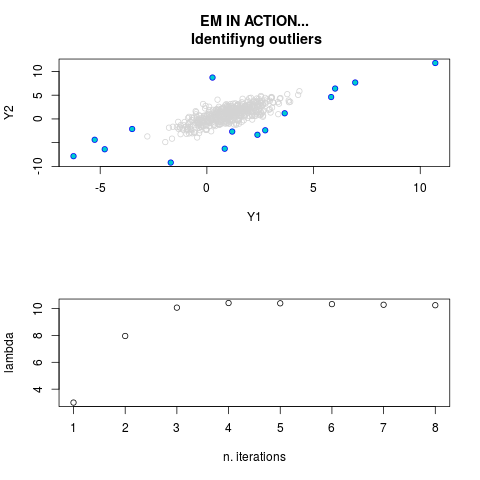
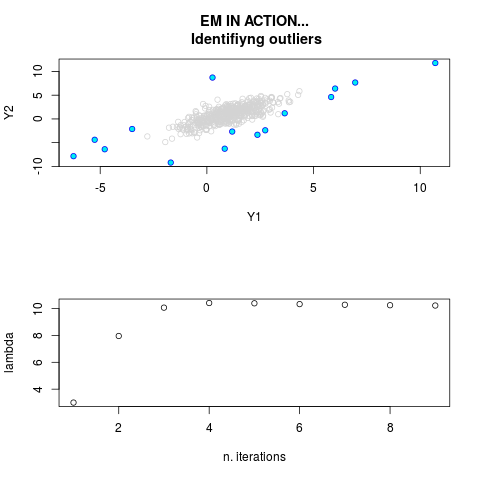
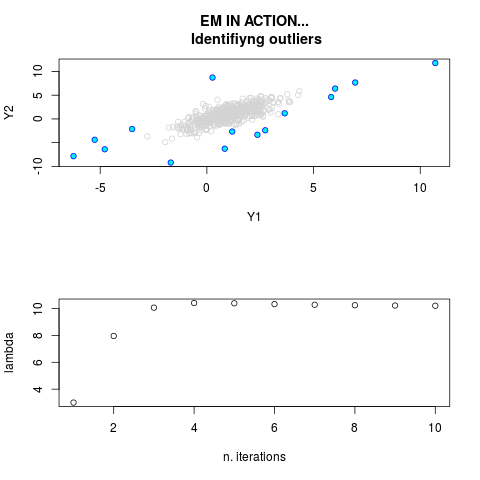
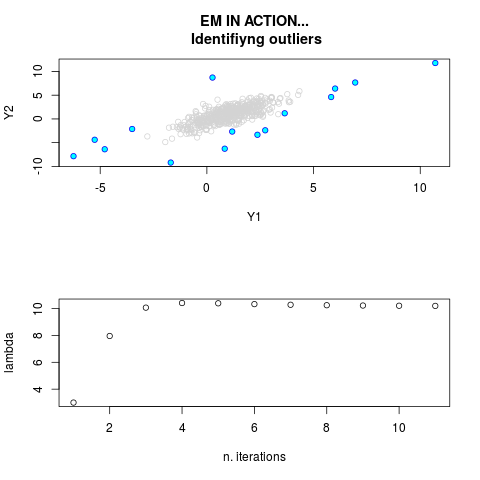
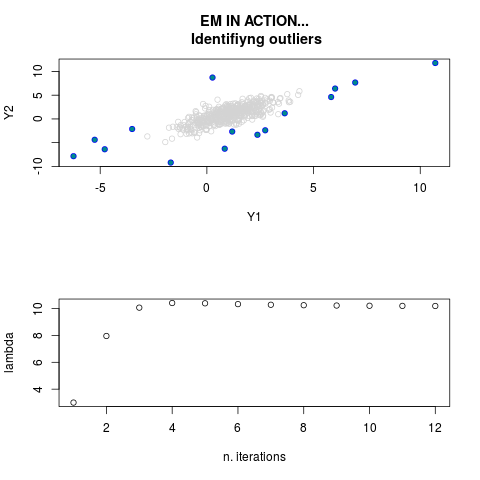
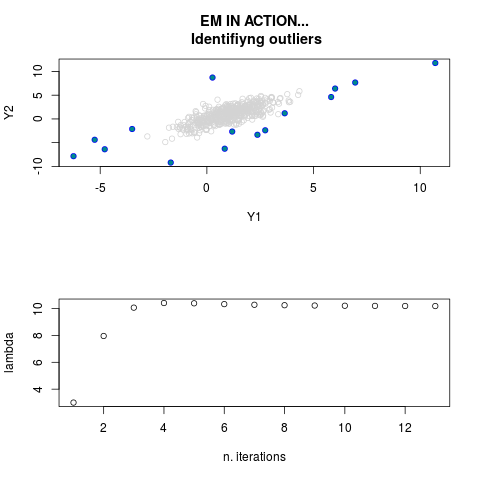
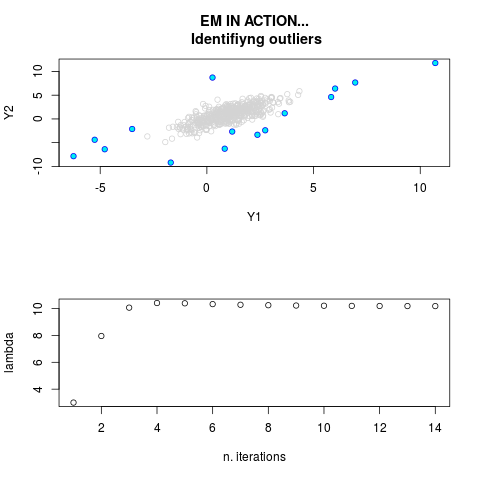
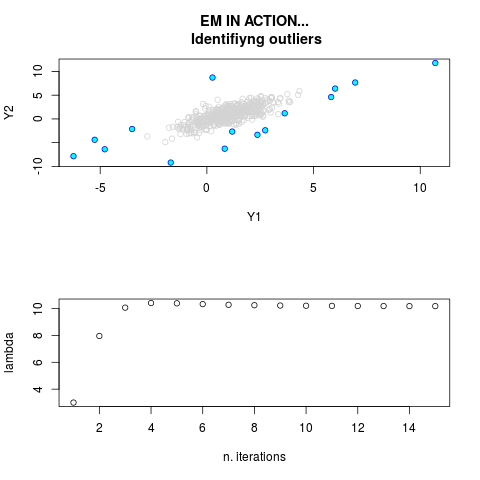
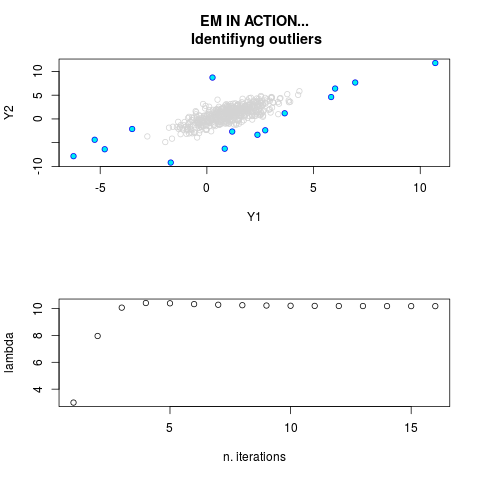
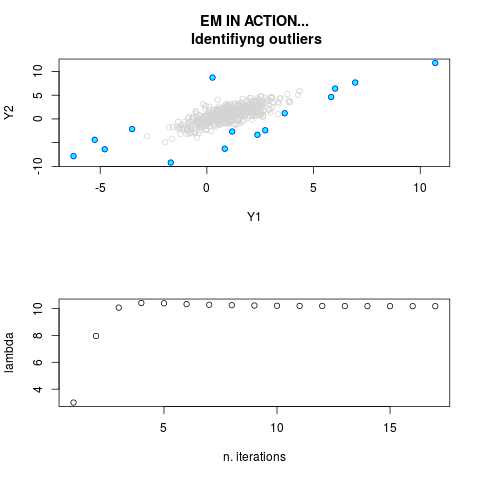
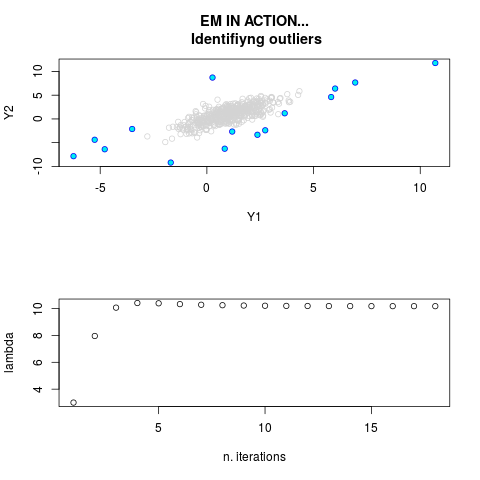
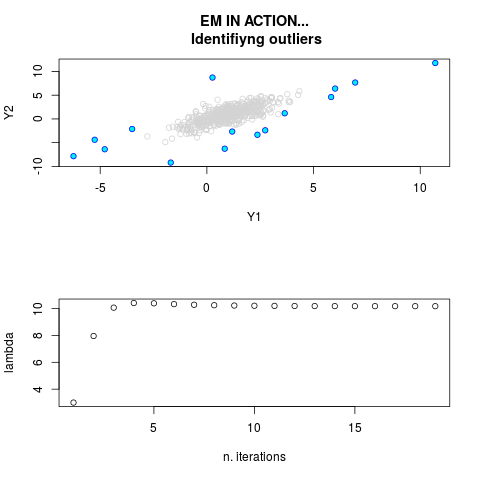
DetailsThis function provides the parameter estimates of a contamination model where a set of According to the estimated model parameters, a matrix of predictions of ‘true’ The model is estimated using complete observations. Missing values in the In case the option ‘model = LN’ is specified, each zero value is changed in 1e-7 and a warning is returned. In order to graphically monitor EM algorithm, a scatter plot is showed where outliers
are depicted as long as they are identified. The trajectory of the Value
Author(s)M. Teresa Buglielli <bugliell@istat.it>, Ugo Guarnera <guarnera@istat.it> ReferencesDi Zio, M., Guarnera, U. (2013) "A Contamination Model for Selective Editing",
Journal of Official Statistics. Volume 29, Issue 4, Pages 539-555 (http://dx.doi.org/10.2478/jos-2013-0039). Buglielli, M.T., Di Zio, M., Guarnera, U. (2010) "Use of Contamination Models for Selective Editing", European Conference on Quality in Survey Statistics Q2010, Helsinki, 4-6 May 2010 Examples
# Parameter estimation with one contaminated variable and one covariate
data(ex1.data)
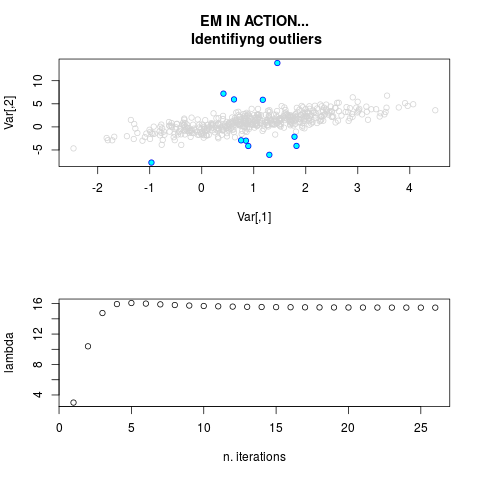
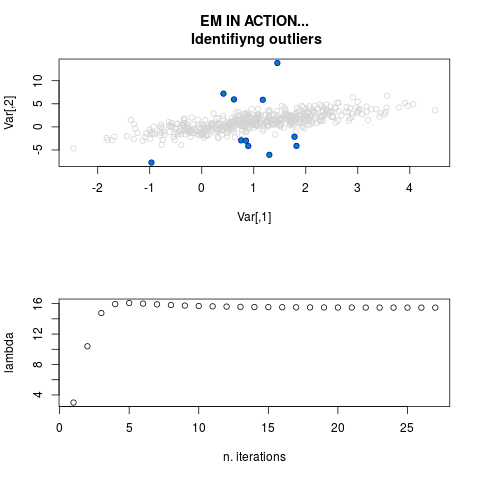
ml.par <- ml.est(y=ex1.data[,"Y1"], x=ex1.data[,"X1"], graph=TRUE)
str(ml.par)
sum(ml.par$outlier) # number of outliers
# Parameter estimation with two contaminated variables and no covariates
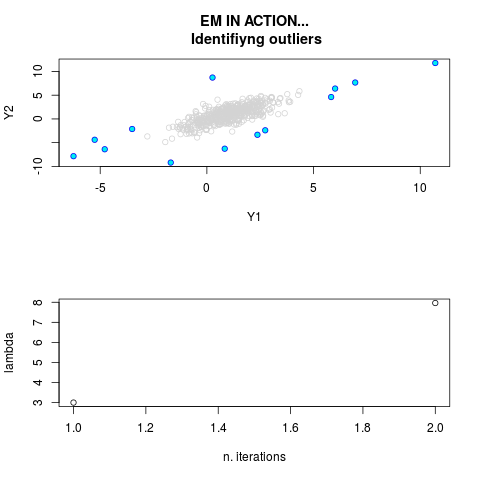
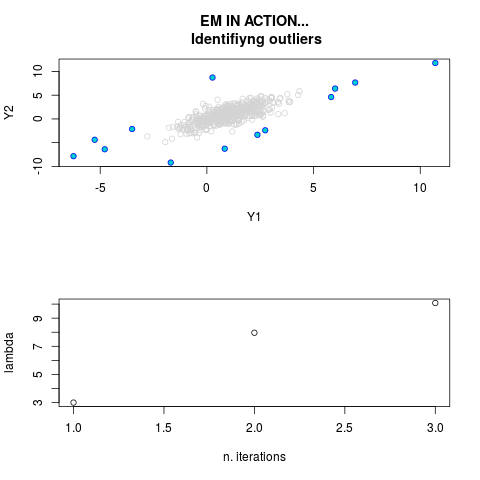
data(ex2.data)
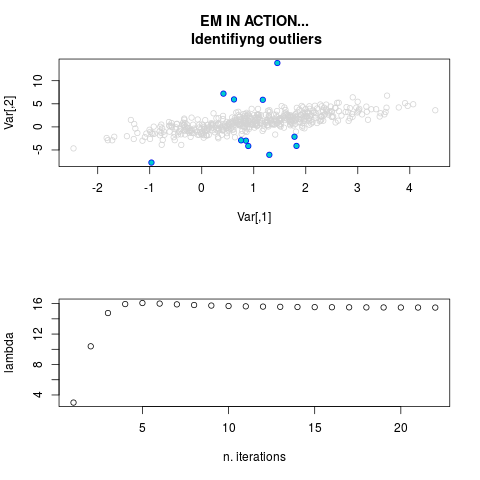
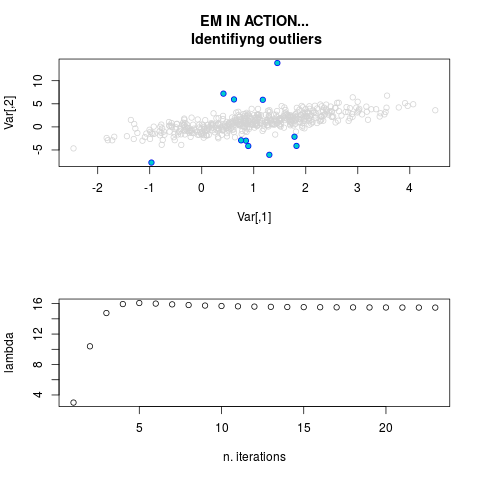
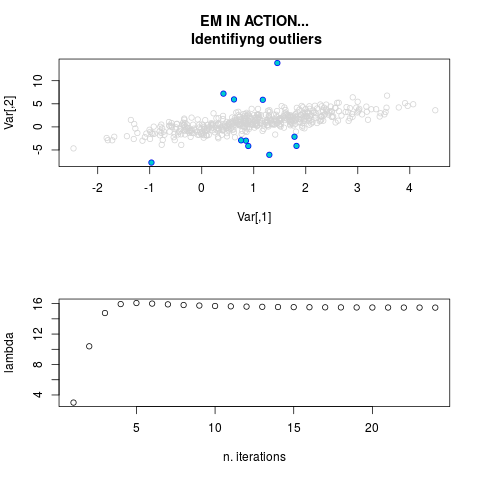
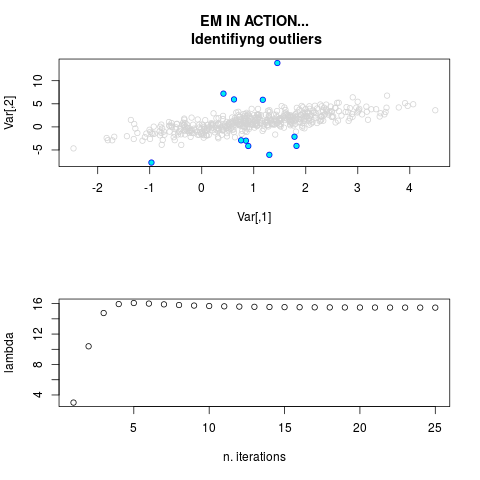
par.joint <- ml.est(y=ex2.data, x=NULL, graph=TRUE)
sum(par.joint$outlier) # number of outliers
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(SeleMix)
Loading required package: mvtnorm
Loading required package: Ecdat
Loading required package: Ecfun
Attaching package: 'Ecfun'
The following object is masked from 'package:base':
sign
Attaching package: 'Ecdat'
The following object is masked from 'package:datasets':
Orange
Loading required package: xtable
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/SeleMix/ml.est.Rd_%03d_medium.png", width=480, height=480)
> ### Name: ml.est
> ### Title: Fitting Contamination Model
> ### Aliases: ml.est
>
> ### ** Examples
>
>
> # Parameter estimation with one contaminated variable and one covariate
> data(ex1.data)
> ml.par <- ml.est(y=ex1.data[,"Y1"], x=ex1.data[,"X1"], graph=TRUE)
> str(ml.par)
List of 11
$ ypred : num [1:500, 1] 1.45 45.62 15.61 41.7 1.04 ...
$ B : num [1:2, 1] -0.152 1.215
$ sigma : num [1, 1] 1.25
$ lambda : num 15.5
$ w : num 0.0479
$ tau : num [1:500] 0.0127 0.0301 0.0122 0.032 0.0234 ...
$ outlier: num [1:500] 0 0 0 0 0 0 0 0 0 0 ...
$ pattern: Factor w/ 1 level "1": 1 1 1 1 1 1 1 1 1 1 ...
$ is.conv: logi TRUE
$ n.iter : num 26
$ bic.aic: Named num [1:4] 1828 1709 911 849
..- attr(*, "names")= chr [1:4] "BIC.norm" "BIC.mix" "AIC.norm" "AIC.mix"
- attr(*, "model")= chr "LN"
- attr(*, "class")= chr [1:2] "list" "mlest"
> sum(ml.par$outlier) # number of outliers
[1] 11
> # Parameter estimation with two contaminated variables and no covariates
> data(ex2.data)
> par.joint <- ml.est(y=ex2.data, x=NULL, graph=TRUE)
> sum(par.joint$outlier) # number of outliers
[1] 15
>
>
>
>
>
> dev.off()
null device
1
>
|