Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Semiparametric Copula Bivariate Regression ModelsDescription
There are many continuous distributions and copula functions that can be employed when using Usage
SemiParBIVProbit(formula, data = list(), weights = NULL, subset = NULL,
Model = "B", BivD = "N",
margins = c("probit","probit"), gamlssfit = FALSE,
fp = FALSE, hess = TRUE, infl.fac = 1,
rinit = 1, rmax = 100,
iterlimsp = 50, tolsp = 1e-07,
gc.l = FALSE, parscale, extra.regI = "t")
Arguments
DetailsThe bivariate models considered in this package consist of two model equations which depend on flexible linear predictors and whose association between the responses is modelled through parameter θ of a standardised bivariate normal distribution or that of a bivariate copula distribution. The linear predictors of the two equations are flexibly specified using parametric components and smooth functions of covariates. The same can be done for the dependence parameter if it makes sense. Estimation is achieved within a penalized likelihood framework with integrated automatic multiple smoothing parameter selection. The use of penalty matrices allows for the suppression of that part of smooth term complexity which has no support from the data. The trade-off between smoothness and fitness is controlled by smoothing parameters associated with the penalty matrices. Smoothing parameters are chosen to minimise an approximate AIC. Details of the underlying fitting methods are given in Radice, Marra and Wojtys (in press). Releases previous to 3.2-7 were based on the algorithms detailed in Marra and Radice (2011, 2013). For sample selection models, if there are factors in the model, before fitting, the user has to ensure that the numbers of factor variables' levels in the selected sample are the same as those in the complete dataset. Even if a model could be fitted in such a situation, the model may produce fits which are not coherent with the nature of the correction sought. As an example consider the situation in which the complete dataset contains a factor variable with five levels and that only three of them appear in the selected sample. For the outcome equation (which is the one of interest) only three levels of such variable exist in the population, but their effects will be corrected for non-random selection using a selection equation in which five levels exist instead. Having differing numbers of factors' levels between complete and selected samples will also make prediction not feasible (an aspect which may be particularly important for selection models); clearly it is not possible to predict the response of interest for the missing entries using a dataset that contains all levels of a factor variable but using an outcome model estimated using a subset of these levels. ValueThe function returns an object of class WARNINGSConvergence failure may sometimes occur. Convergence can be checked using In such a situation, the user may use some extra regularisation (see The above suggestions may help, especially the latter option. However, the user should also consider re-specifying the model and/or using a diferrent dependence structure and/or checking that the chosen marginal distribution fit the responses well. In our experience, we found that convergence failure typically occurs when the model has been misspecified and/or the sample size (and/or number of selected observations in selection models) is low compared to the complexity of the model. Examples of misspecification include using a Clayton copula rotated by 90 degrees when a positive association between the margins is present instead, using marginal distributions that do not fit the responses (again, this is a bit more relevant when one of the two responses is continuous), and employing a copula which does not accommodate the type and/or strength of the dependence between the margins (e.g., using AMH when the association between the margins is strong). It is also worth bearing in mind that the use of a three parameter marginal distribution requires the data to be more informative than a situation in which a two parameter distribution is used instead. When comparing competing models (for instance, by keeping the linear predictor specifications fixed and changing the copula), if the computing time for a set of alternatives is considerably higher than that of another set then it may mean that the more computationally demanding models are not able to fit the data very well (as a higher number of iterations is required to reach convergence). As a practical check, this may be verified by fitting all competing models and, provided convergence is achieved, comparing their respective AIC and BICs, for instance. In the contexts of endogeneity and non-random sample selection, extra attention is required when specifying the dependence parameter as a function of covariates. This is because in these situations the dependence parameter mainly models the association between the unobserved confounders in the two equations. Therefore, this option would make sense when it is believed that the strength of the association between the unobservables in the two equations varies based on some grouping factor or across geographical areas, for instance. Author(s)Maintainer: Giampiero Marra giampiero.marra@ucl.ac.uk ReferencesMarra G. and Radice R. (2011), Estimation of a Semiparametric Recursive Bivariate Probit in the Presence of Endogeneity. Canadian Journal of Statistics, 39(2), 259-279. Marra G. and Radice R. (2013), A Penalized Likelihood Estimation Approach to Semiparametric Sample Selection Binary Response Modeling. Electronic Journal of Statistics, 7, 1432-1455. Marra G., Radice R. and Missiroli S. (2014), Testing the Hypothesis of Absence of Unobserved Confounding in Semiparametric Bivariate Probit Models. Computational Statistics, 29(3-4), 715-741. Marra G., Radice R. and Filippou P. (in press), Regression Spline Bivariate Probit Models: A Practical Approach to Testing for Exogeneity. Communications in Statistics - Simulation and Computation. Marra G., Radice R., Barnighausen T., Wood S.N. and McGovern M.E. (submitted). A Simultaneous Equation Approach to Estimating HIV Prevalence with Non-Ignorable Missing Responses. McGovern M.E., Barnighausen T., Marra G. and Radice R. (2015), On the Assumption of Joint Normality in Selection Models: A Copula Approach Applied to Estimating HIV Prevalence. Epidemiology, 26(2), 229-237. Radice R., Marra G. and M. Wojtys (in press), Copula Regression Spline Models for Binary Outcomes. Statistics and Computing. Marra G. and Wood S.N. (2011), Practical Variable Selection for Generalized Additive Models. Computational Statistics and Data Analysis, 55(7), 2372-2387. Marra G. and Wood S.N. (2012), Coverage Properties of Confidence Intervals for Generalized Additive Model Components. Scandinavian Journal of Statistics, 39(1), 53-74. Poirier D.J. (1980), Partial Observability in Bivariate Probit Models. Journal of Econometrics, 12, 209-217. Wood S.N. (2004), Stable and efficient multiple smoothing parameter estimation for generalized additive models. Journal of the American Statistical Association, 99(467), 673-686. See Also
Examples
library(SemiParBIVProbit)
############
## EXAMPLE 1
############
## Generate data
## Correlation between the two equations 0.5 - Sample size 400
set.seed(0)
n <- 400
Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
u <- rMVN(n, rep(0,2), Sigma)
x1 <- round(runif(n)); x2 <- runif(n); x3 <- runif(n)
f1 <- function(x) cos(pi*2*x) + sin(pi*x)
f2 <- function(x) x+exp(-30*(x-0.5)^2)
y1 <- ifelse(-1.55 + 2*x1 + f1(x2) + u[,1] > 0, 1, 0)
y2 <- ifelse(-0.25 - 1.25*x1 + f2(x2) + u[,2] > 0, 1, 0)
dataSim <- data.frame(y1, y2, x1, x2, x3)
#
#
## CLASSIC BIVARIATE PROBIT
out <- SemiParBIVProbit(list(y1 ~ x1 + x2 + x3,
y2 ~ x1 + x2 + x3),
data = dataSim)
conv.check(out)
summary(out)
AIC(out)
BIC(out)
## SEMIPARAMETRIC BIVARIATE PROBIT
## "cr" cubic regression spline basis - "cs" shrinkage version of "cr"
## "tp" thin plate regression spline basis - "ts" shrinkage version of "tp"
## for smooths of one variable, "cr/cs" and "tp/ts" achieve similar results
## k is the basis dimension - default is 10
## m is the order of the penalty for the specific term - default is 2
## For COPULA models use BivD argument
out <- SemiParBIVProbit(list(y1 ~ x1 + s(x2, bs = "tp", k = 10, m = 2) + s(x3),
y2 ~ x1 + s(x2) + s(x3)),
data = dataSim)
conv.check(out)
summary(out, cm.plot = TRUE)
AIC(out)
## estimated smooth function plots - red lines are true curves
x2 <- sort(x2)
f1.x2 <- f1(x2)[order(x2)] - mean(f1(x2))
f2.x2 <- f2(x2)[order(x2)] - mean(f2(x2))
f3.x3 <- rep(0, length(x3))
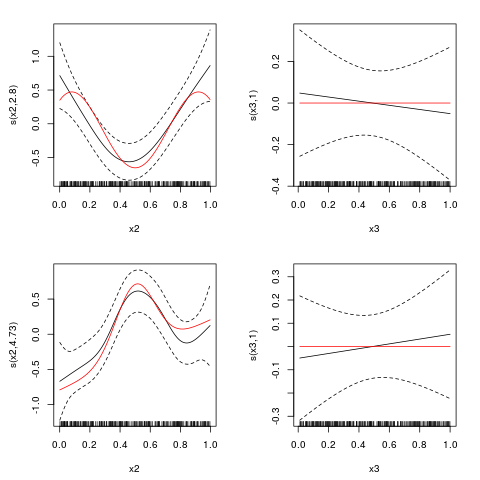
par(mfrow=c(2,2),mar=c(4.5,4.5,2,2))
plot(out, eq = 1, select = 1, seWithMean = TRUE, scale = 0)
lines(x2, f1.x2, col = "red")
plot(out, eq = 1, select = 2, seWithMean = TRUE, scale = 0)
lines(x3, f3.x3, col = "red")
plot(out, eq = 2, select = 1, seWithMean = TRUE, scale = 0)
lines(x2, f2.x2, col = "red")
plot(out, eq = 2, select = 2, seWithMean = TRUE, scale = 0)
lines(x3, f3.x3, col = "red")
## p-values suggest to drop x3 from both equations, with a stronger
## evidence for eq. 2. This can be also achieved using shrinkage smoothers
outSS <- SemiParBIVProbit(list(y1 ~ x1 + s(x2, bs = "ts") + s(x3, bs = "cs"),
y2 ~ x1 + s(x2, bs = "cs") + s(x3, bs = "ts")),
data = dataSim)
conv.check(outSS)
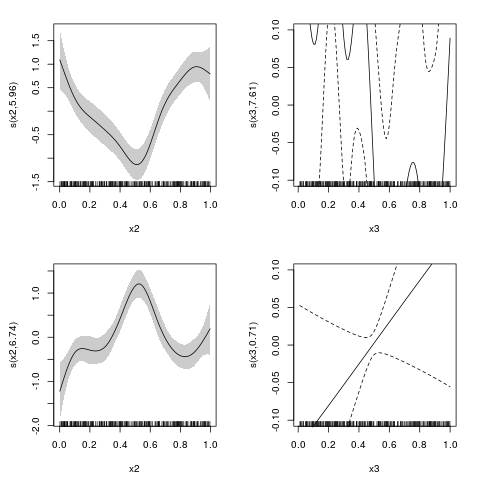
plot(outSS, eq = 1, select = 1, scale = 0, shade = TRUE)
plot(outSS, eq = 1, select = 2, ylim = c(-0.1,0.1))
plot(outSS, eq = 2, select = 1, scale = 0, shade = TRUE)
plot(outSS, eq = 2, select = 2, ylim = c(-0.1,0.1))
## Not run:
## SEMIPARAMETRIC BIVARIATE PROBIT with association parameter
## depending on covariates as well
eq.mu.1 <- y1 ~ x1 + s(x2)
eq.mu.2 <- y2 ~ x1 + s(x2)
eq.theta <- ~ x1 + s(x2)
fl <- list(eq.mu.1, eq.mu.2, eq.theta)
outD <- SemiParBIVProbit(fl, data = dataSim)
conv.check(outD)
summary(outD)
outD$theta
plot(outD, eq = 1, seWithMean = TRUE)
plot(outD, eq = 2, seWithMean = TRUE)
plot(outD, eq = 3, seWithMean = TRUE)
graphics.off()
#
#
############
## EXAMPLE 2
############
## Generate data with one endogenous variable
## and exclusion restriction
set.seed(0)
n <- 400
Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
u <- rMVN(n, rep(0,2), Sigma)
cov <- rMVN(n, rep(0,2), Sigma)
cov <- pnorm(cov)
x1 <- round(cov[,1]); x2 <- cov[,2]
f1 <- function(x) cos(pi*2*x) + sin(pi*x)
f2 <- function(x) x+exp(-30*(x-0.5)^2)
y1 <- ifelse(-1.55 + 2*x1 + f1(x2) + u[,1] > 0, 1, 0)
y2 <- ifelse(-0.25 - 1.25*y1 + f2(x2) + u[,2] > 0, 1, 0)
dataSim <- data.frame(y1, y2, x1, x2)
#
## Testing the hypothesis of absence of endogeneity...
LM.bpm(list(y1 ~ x1 + s(x2), y2 ~ y1 + s(x2)), dataSim, Model = "B")
# p-value suggests presence of endogeneity, hence fit a bivariate model
## CLASSIC RECURSIVE BIVARIATE PROBIT
out <- SemiParBIVProbit(list(y1 ~ x1 + x2,
y2 ~ y1 + x2),
data = dataSim)
conv.check(out)
summary(out)
AIC(out); BIC(out)
## SEMIPARAMETRIC RECURSIVE BIVARIATE PROBIT
out <- SemiParBIVProbit(list(y1 ~ x1 + s(x2),
y2 ~ y1 + s(x2)),
data = dataSim)
conv.check(out)
summary(out)
AIC(out); BIC(out)
#
## Testing the hypothesis of absence of endogeneity post estimation...
gt.bpm(out)
#
## reatment effect, risk ratio and odds ratio with CIs
mb(y1, y2, Model = "B")
AT(out, nm.end = "y1", hd.plot = TRUE)
RR(out, nm.end = "y1")
OR(out, nm.end = "y1")
AT(out, nm.end = "y1", type = "univariate")
## try a Clayton copula model...
outC <- SemiParBIVProbit(list(y1 ~ x1 + s(x2),
y2 ~ y1 + s(x2)),
data = dataSim, BivD = "C0")
conv.check(outC)
summary(outC)
AT(outC, nm.end = "y1")
## try a Joe copula model...
outJ <- SemiParBIVProbit(list(y1 ~ x1 + s(x2),
y2 ~ y1 + s(x2)),
data = dataSim, BivD = "J0")
conv.check(outJ)
summary(outJ, cm.plot = TRUE)
AT(outJ, "y1")
VuongClarke(out, outJ)
#
## recursive bivariate probit modelling with unpenalized splines
## can be achieved as follows
outFP <- SemiParBIVProbit(list(y1 ~ x1 + s(x2, bs = "cr", k = 5),
y2 ~ y1 + s(x2, bs = "cr", k = 6)),
fp = TRUE, data = dataSim)
conv.check(outFP)
summary(outFP)
# in the above examples a third equation could be introduced
# as illustrated in Example 1
#
#################
## See also ?meps
#################
############
## EXAMPLE 3
############
## Generate data with a non-random sample selection mechanism
## and exclusion restriction
set.seed(0)
n <- 2000
Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
u <- rMVN(n, rep(0,2), Sigma)
SigmaC <- matrix(0.5, 3, 3); diag(SigmaC) <- 1
cov <- rMVN(n, rep(0,3), SigmaC)
cov <- pnorm(cov)
bi <- round(cov[,1]); x1 <- cov[,2]; x2 <- cov[,3]
f11 <- function(x) -0.7*(4*x + 2.5*x^2 + 0.7*sin(5*x) + cos(7.5*x))
f12 <- function(x) -0.4*( -0.3 - 1.6*x + sin(5*x))
f21 <- function(x) 0.6*(exp(x) + sin(2.9*x))
ys <- 0.58 + 2.5*bi + f11(x1) + f12(x2) + u[, 1] > 0
y <- -0.68 - 1.5*bi + f21(x1) + + u[, 2] > 0
yo <- y*(ys > 0)
dataSim <- data.frame(y, ys, yo, bi, x1, x2)
## Testing the hypothesis of absence of non-random sample selection...
LM.bpm(list(ys ~ bi + s(x1) + s(x2), yo ~ bi + s(x1)), dataSim, Model = "BSS")
# p-value suggests presence of sample selection, hence fit a bivariate model
#
## SEMIPARAMETRIC SAMPLE SELECTION BIVARIATE PROBIT
## the first equation MUST be the selection equation
out <- SemiParBIVProbit(list(ys ~ bi + s(x1) + s(x2),
yo ~ bi + s(x1)),
data = dataSim, Model = "BSS")
conv.check(out)
gt.bpm(out)
## compare the two summary outputs
## the second output produces a summary of the results obtained when
## selection bias is not accounted for
summary(out)
summary(out$gam2)
## corrected predicted probability that 'yo' is equal to 1
mb(ys, yo, Model = "BSS")
prev(out, hd.plot = TRUE)
prev(out, type = "univariate", hd.plot = TRUE)
## estimated smooth function plots
## the red line is the true curve
## the blue line is the univariate model curve not accounting for selection bias
x1.s <- sort(x1[dataSim$ys>0])
f21.x1 <- f21(x1.s)[order(x1.s)]-mean(f21(x1.s))
plot(out, eq = 2, ylim = c(-1.65,0.95)); lines(x1.s, f21.x1, col="red")
par(new = TRUE)
plot(out$gam2, se = FALSE, col = "blue", ylim = c(-1.65,0.95),
ylab = "", rug = FALSE)
#
#
## try a Clayton copula model...
outC <- SemiParBIVProbit(list(ys ~ bi + s(x1) + s(x2),
yo ~ bi + s(x1)),
data = dataSim, Model = "BSS", BivD = "C0")
conv.check(outC)
summary(outC, cm.plot = TRUE)
prev(outC)
# in the above examples a third equation could be introduced
# as illustrated in Example 1
#
################
## See also ?hiv
################
############
## EXAMPLE 4
############
## Generate data with partial observability
set.seed(0)
n <- 10000
Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
u <- rMVN(n, rep(0,2), Sigma)
x1 <- round(runif(n)); x2 <- runif(n); x3 <- runif(n)
y1 <- ifelse(-1.55 + 2*x1 + x2 + u[,1] > 0, 1, 0)
y2 <- ifelse( 0.45 - x3 + u[,2] > 0, 1, 0)
y <- y1*y2
dataSim <- data.frame(y, x1, x2, x3)
## BIVARIATE PROBIT with Partial Observability
out <- SemiParBIVProbit(list(y ~ x1 + x2,
y ~ x3),
data = dataSim, Model = "BPO")
conv.check(out)
summary(out)
# first ten estimated probabilities for the four events from object out
cbind(out$p11, out$p10, out$p00, out$p01)[1:10,]
# case with smooth function
# (more computationally intensive)
f1 <- function(x) cos(pi*2*x) + sin(pi*x)
y1 <- ifelse(-1.55 + 2*x1 + f1(x2) + u[,1] > 0, 1, 0)
y2 <- ifelse( 0.45 - x3 + u[,2] > 0, 1, 0)
y <- y1*y2
dataSim <- data.frame(y, x1, x2, x3)
out <- SemiParBIVProbit(list(y ~ x1 + s(x2),
y ~ x3),
data = dataSim, Model = "BPO")
conv.check(out)
summary(out, cm.plot = TRUE)
# plot estimated and true functions
x2 <- sort(x2); f1.x2 <- f1(x2)[order(x2)] - mean(f1(x2))
plot(out, eq = 1, scale = 0); lines(x2, f1.x2, col = "red")
#
################
## See also ?war
################
############
## EXAMPLE 5
############
## Generate data with one endogenous binary variable
## and continuous outcome
set.seed(0)
n <- 1000
Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
u <- rMVN(n, rep(0,2), Sigma)
cov <- rMVN(n, rep(0,2), Sigma)
cov <- pnorm(cov)
x1 <- round(cov[,1]); x2 <- cov[,2]
f1 <- function(x) cos(pi*2*x) + sin(pi*x)
f2 <- function(x) x+exp(-30*(x-0.5)^2)
y1 <- ifelse(-1.55 + 2*x1 + f1(x2) + u[,1] > 0, 1, 0)
y2 <- -0.25 - 1.25*y1 + f2(x2) + u[,2]
dataSim <- data.frame(y1, y2, x1, x2)
## RECURSIVE Model
rc <- resp.check(y2, margin = "N", print.par = TRUE, loglik = TRUE)
AIC(rc); BIC(rc)
out <- SemiParBIVProbit(list(y1 ~ x1 + x2,
y2 ~ y1 + x2),
data = dataSim, margins = c("probit","N"))
conv.check(out)
summary(out)
post.check(out)
## SEMIPARAMETRIC RECURSIVE Model
eq.mu.1 <- y1 ~ x1 + s(x2)
eq.mu.2 <- y2 ~ y1 + s(x2)
eq.sigma2 <- ~ 1
eq.theta <- ~ 1
fl <- list(eq.mu.1, eq.mu.2, eq.sigma2, eq.theta)
out <- SemiParBIVProbit(fl, data = dataSim,
margins = c("probit","N"), gamlssfit = TRUE)
conv.check(out)
summary(out)
post.check(out)
jc.probs(out, 1, 1.5, intervals = TRUE)[1:4,]
AT(out, nm.end = "y1")
AT(out, nm.end = "y1", type = "univariate")
#
#
############
## EXAMPLE 6
############
## Generate data with one endogenous continuous exposure
## and binary outcome
set.seed(0)
n <- 1000
Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
u <- rMVN(n, rep(0,2), Sigma)
cov <- rMVN(n, rep(0,2), Sigma)
cov <- pnorm(cov)
x1 <- round(cov[,1]); x2 <- cov[,2]
f1 <- function(x) cos(pi*2*x) + sin(pi*x)
f2 <- function(x) x+exp(-30*(x-0.5)^2)
y1 <- -0.25 - 2*x1 + f2(x2) + u[,2]
y2 <- ifelse(-0.25 - 0.25*y1 + f1(x2) + u[,1] > 0, 1, 0)
dataSim <- data.frame(y1, y2, x1, x2)
eq.mu.1 <- y2 ~ y1 + s(x2)
eq.mu.2 <- y1 ~ x1 + s(x2)
eq.sigma2 <- ~ 1
eq.theta <- ~ 1
fl <- list(eq.mu.1, eq.mu.2, eq.sigma2, eq.theta)
out <- SemiParBIVProbit(fl, data = dataSim,
margins = c("probit","N"))
conv.check(out)
summary(out)
post.check(out)
AT(out, nm.end = "y1")
AT(out, nm.end = "y1", type = "univariate")
RR(out, nm.end = "y1", rr.plot = TRUE)
RR(out, nm.end = "y1", type = "univariate")
OR(out, nm.end = "y1", or.plot = TRUE)
OR(out, nm.end = "y1", type = "univariate")
## End(Not run)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(SemiParBIVProbit)
Loading required package: mgcv
Loading required package: nlme
This is mgcv 1.8-12. For overview type 'help("mgcv-package")'.
This is SemiParBIVProbit 3.7-1.
For overview type 'help("SemiParBIVProbit-package")'.
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/SemiParBIVProbit/SemiParBIVProbit.Rd_%03d_medium.png", width=480, height=480)
> ### Name: SemiParBIVProbit
> ### Title: Semiparametric Copula Bivariate Regression Models
> ### Aliases: SemiParBIVProbit
> ### Keywords: non-random sample selection semiparametric bivariate
> ### modelling partial observability endogeneity smooth regression spline
> ### shrinkage smoother variable selection
>
> ### ** Examples
>
>
> library(SemiParBIVProbit)
>
> ############
> ## EXAMPLE 1
> ############
> ## Generate data
> ## Correlation between the two equations 0.5 - Sample size 400
>
> set.seed(0)
>
> n <- 400
>
> Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
> u <- rMVN(n, rep(0,2), Sigma)
>
> x1 <- round(runif(n)); x2 <- runif(n); x3 <- runif(n)
>
> f1 <- function(x) cos(pi*2*x) + sin(pi*x)
> f2 <- function(x) x+exp(-30*(x-0.5)^2)
>
> y1 <- ifelse(-1.55 + 2*x1 + f1(x2) + u[,1] > 0, 1, 0)
> y2 <- ifelse(-0.25 - 1.25*x1 + f2(x2) + u[,2] > 0, 1, 0)
>
> dataSim <- data.frame(y1, y2, x1, x2, x3)
>
> #
> #
>
> ## CLASSIC BIVARIATE PROBIT
>
> out <- SemiParBIVProbit(list(y1 ~ x1 + x2 + x3,
+ y2 ~ x1 + x2 + x3),
+ data = dataSim)
> conv.check(out)
Largest absolute gradient value: 9.321016e-14
Observed information matrix is positive definite
Eigenvalue range: [9.528687,419.9106]
Trust region iterations: 3
> summary(out)
COPULA: Gaussian
MARGIN 1: Bernoulli
MARGIN 2: Bernoulli
EQUATION 1
Link function for mu.1: probit
Formula: y1 ~ x1 + x2 + x3
Parametric coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.94936 0.20532 -4.624 3.77e-06 ***
x1 1.97383 0.15150 13.028 < 2e-16 ***
x2 0.20574 0.25474 0.808 0.419
x3 -0.06393 0.25939 -0.246 0.805
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
EQUATION 2
Link function for mu.2: probit
Formula: y2 ~ x1 + x2 + x3
Parametric coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.04474 0.17736 0.252 0.80084
x1 -1.07918 0.13393 -8.058 7.76e-16 ***
x2 0.66869 0.23016 2.905 0.00367 **
x3 0.08599 0.23228 0.370 0.71124
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
n = 400 theta = 0.282(0.062,0.458) tau = 0.182(0.0395,0.303)
total edf = 9
> AIC(out)
[1] 840.3036
> BIC(out)
[1] 876.2268
>
> ## SEMIPARAMETRIC BIVARIATE PROBIT
>
> ## "cr" cubic regression spline basis - "cs" shrinkage version of "cr"
> ## "tp" thin plate regression spline basis - "ts" shrinkage version of "tp"
> ## for smooths of one variable, "cr/cs" and "tp/ts" achieve similar results
> ## k is the basis dimension - default is 10
> ## m is the order of the penalty for the specific term - default is 2
> ## For COPULA models use BivD argument
>
> out <- SemiParBIVProbit(list(y1 ~ x1 + s(x2, bs = "tp", k = 10, m = 2) + s(x3),
+ y2 ~ x1 + s(x2) + s(x3)),
+ data = dataSim)
> conv.check(out)
Largest absolute gradient value: 1.485398e-09
Observed information matrix is positive definite
Eigenvalue range: [0.2372136,5.216988e+13]
Trust region iterations before smoothing parameter estimation: 4
Loops for smoothing parameter estimation: 3
Trust region iterations within smoothing loops: 4
> summary(out, cm.plot = TRUE)
COPULA: Gaussian
MARGIN 1: Bernoulli
MARGIN 2: Bernoulli
EQUATION 1
Link function for mu.1: probit
Formula: y1 ~ x1 + s(x2, bs = "tp", k = 10, m = 2) + s(x3)
Parametric coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.9852 0.1094 -9.005 <2e-16 ***
x1 2.1885 0.1691 12.940 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Smooth components' approximate significance:
edf Ref.df Chi.sq p-value
s(x2) 2.805 3.499 31.721 1.87e-06 ***
s(x3) 1.000 1.000 0.134 0.714
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
EQUATION 2
Link function for mu.2: probit
Formula: y2 ~ x1 + s(x2) + s(x3)
Parametric coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.43374 0.09316 4.656 3.22e-06 ***
x1 -1.14477 0.13880 -8.248 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Smooth components' approximate significance:
edf Ref.df Chi.sq p-value
s(x2) 4.728 5.784 33.810 6.81e-06 ***
s(x3) 1.000 1.000 0.189 0.664
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
n = 400 theta = 0.484(0.324,0.646) tau = 0.322(0.21,0.447)
total edf = 14.5
> AIC(out)
[1] 774.3511
>
>
> ## estimated smooth function plots - red lines are true curves
>
> x2 <- sort(x2)
> f1.x2 <- f1(x2)[order(x2)] - mean(f1(x2))
> f2.x2 <- f2(x2)[order(x2)] - mean(f2(x2))
> f3.x3 <- rep(0, length(x3))
>
> par(mfrow=c(2,2),mar=c(4.5,4.5,2,2))
> plot(out, eq = 1, select = 1, seWithMean = TRUE, scale = 0)
> lines(x2, f1.x2, col = "red")
> plot(out, eq = 1, select = 2, seWithMean = TRUE, scale = 0)
> lines(x3, f3.x3, col = "red")
> plot(out, eq = 2, select = 1, seWithMean = TRUE, scale = 0)
> lines(x2, f2.x2, col = "red")
> plot(out, eq = 2, select = 2, seWithMean = TRUE, scale = 0)
> lines(x3, f3.x3, col = "red")
>
> ## p-values suggest to drop x3 from both equations, with a stronger
> ## evidence for eq. 2. This can be also achieved using shrinkage smoothers
>
> outSS <- SemiParBIVProbit(list(y1 ~ x1 + s(x2, bs = "ts") + s(x3, bs = "cs"),
+ y2 ~ x1 + s(x2, bs = "cs") + s(x3, bs = "ts")),
+ data = dataSim)
Warning message:
Largest absolute gradient value is not close to 0.
Read the WARNINGS section in ?SemiParBIVProbit.
> conv.check(outSS)
Largest absolute gradient value: 1.747239
Observed information matrix is positive definite
Eigenvalue range: [5.711081e-129,449317]
Trust region iterations before smoothing parameter estimation: 4
Loops for smoothing parameter estimation: 4
Trust region iterations within smoothing loops: 174
>
> plot(outSS, eq = 1, select = 1, scale = 0, shade = TRUE)
> plot(outSS, eq = 1, select = 2, ylim = c(-0.1,0.1))
> plot(outSS, eq = 2, select = 1, scale = 0, shade = TRUE)
> plot(outSS, eq = 2, select = 2, ylim = c(-0.1,0.1))
>
> ## Not run:
> ##D
> ##D
> ##D ## SEMIPARAMETRIC BIVARIATE PROBIT with association parameter
> ##D ## depending on covariates as well
> ##D
> ##D eq.mu.1 <- y1 ~ x1 + s(x2)
> ##D eq.mu.2 <- y2 ~ x1 + s(x2)
> ##D eq.theta <- ~ x1 + s(x2)
> ##D
> ##D fl <- list(eq.mu.1, eq.mu.2, eq.theta)
> ##D
> ##D outD <- SemiParBIVProbit(fl, data = dataSim)
> ##D conv.check(outD)
> ##D summary(outD)
> ##D outD$theta
> ##D
> ##D plot(outD, eq = 1, seWithMean = TRUE)
> ##D plot(outD, eq = 2, seWithMean = TRUE)
> ##D plot(outD, eq = 3, seWithMean = TRUE)
> ##D graphics.off()
> ##D
> ##D #
> ##D #
> ##D
> ##D ############
> ##D ## EXAMPLE 2
> ##D ############
> ##D ## Generate data with one endogenous variable
> ##D ## and exclusion restriction
> ##D
> ##D set.seed(0)
> ##D
> ##D n <- 400
> ##D
> ##D Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
> ##D u <- rMVN(n, rep(0,2), Sigma)
> ##D
> ##D cov <- rMVN(n, rep(0,2), Sigma)
> ##D cov <- pnorm(cov)
> ##D x1 <- round(cov[,1]); x2 <- cov[,2]
> ##D
> ##D f1 <- function(x) cos(pi*2*x) + sin(pi*x)
> ##D f2 <- function(x) x+exp(-30*(x-0.5)^2)
> ##D
> ##D y1 <- ifelse(-1.55 + 2*x1 + f1(x2) + u[,1] > 0, 1, 0)
> ##D y2 <- ifelse(-0.25 - 1.25*y1 + f2(x2) + u[,2] > 0, 1, 0)
> ##D
> ##D dataSim <- data.frame(y1, y2, x1, x2)
> ##D
> ##D #
> ##D
> ##D ## Testing the hypothesis of absence of endogeneity...
> ##D
> ##D LM.bpm(list(y1 ~ x1 + s(x2), y2 ~ y1 + s(x2)), dataSim, Model = "B")
> ##D
> ##D # p-value suggests presence of endogeneity, hence fit a bivariate model
> ##D
> ##D
> ##D ## CLASSIC RECURSIVE BIVARIATE PROBIT
> ##D
> ##D out <- SemiParBIVProbit(list(y1 ~ x1 + x2,
> ##D y2 ~ y1 + x2),
> ##D data = dataSim)
> ##D conv.check(out)
> ##D summary(out)
> ##D AIC(out); BIC(out)
> ##D
> ##D ## SEMIPARAMETRIC RECURSIVE BIVARIATE PROBIT
> ##D
> ##D out <- SemiParBIVProbit(list(y1 ~ x1 + s(x2),
> ##D y2 ~ y1 + s(x2)),
> ##D data = dataSim)
> ##D conv.check(out)
> ##D summary(out)
> ##D AIC(out); BIC(out)
> ##D
> ##D #
> ##D
> ##D ## Testing the hypothesis of absence of endogeneity post estimation...
> ##D
> ##D gt.bpm(out)
> ##D
> ##D #
> ##D ## reatment effect, risk ratio and odds ratio with CIs
> ##D
> ##D mb(y1, y2, Model = "B")
> ##D AT(out, nm.end = "y1", hd.plot = TRUE)
> ##D RR(out, nm.end = "y1")
> ##D OR(out, nm.end = "y1")
> ##D AT(out, nm.end = "y1", type = "univariate")
> ##D
> ##D
> ##D ## try a Clayton copula model...
> ##D
> ##D outC <- SemiParBIVProbit(list(y1 ~ x1 + s(x2),
> ##D y2 ~ y1 + s(x2)),
> ##D data = dataSim, BivD = "C0")
> ##D conv.check(outC)
> ##D summary(outC)
> ##D AT(outC, nm.end = "y1")
> ##D
> ##D ## try a Joe copula model...
> ##D
> ##D outJ <- SemiParBIVProbit(list(y1 ~ x1 + s(x2),
> ##D y2 ~ y1 + s(x2)),
> ##D data = dataSim, BivD = "J0")
> ##D conv.check(outJ)
> ##D summary(outJ, cm.plot = TRUE)
> ##D AT(outJ, "y1")
> ##D
> ##D
> ##D VuongClarke(out, outJ)
> ##D
> ##D #
> ##D ## recursive bivariate probit modelling with unpenalized splines
> ##D ## can be achieved as follows
> ##D
> ##D outFP <- SemiParBIVProbit(list(y1 ~ x1 + s(x2, bs = "cr", k = 5),
> ##D y2 ~ y1 + s(x2, bs = "cr", k = 6)),
> ##D fp = TRUE, data = dataSim)
> ##D conv.check(outFP)
> ##D summary(outFP)
> ##D
> ##D # in the above examples a third equation could be introduced
> ##D # as illustrated in Example 1
> ##D
> ##D #
> ##D #################
> ##D ## See also ?meps
> ##D #################
> ##D
> ##D ############
> ##D ## EXAMPLE 3
> ##D ############
> ##D ## Generate data with a non-random sample selection mechanism
> ##D ## and exclusion restriction
> ##D
> ##D set.seed(0)
> ##D
> ##D n <- 2000
> ##D
> ##D Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
> ##D u <- rMVN(n, rep(0,2), Sigma)
> ##D
> ##D SigmaC <- matrix(0.5, 3, 3); diag(SigmaC) <- 1
> ##D cov <- rMVN(n, rep(0,3), SigmaC)
> ##D cov <- pnorm(cov)
> ##D bi <- round(cov[,1]); x1 <- cov[,2]; x2 <- cov[,3]
> ##D
> ##D f11 <- function(x) -0.7*(4*x + 2.5*x^2 + 0.7*sin(5*x) + cos(7.5*x))
> ##D f12 <- function(x) -0.4*( -0.3 - 1.6*x + sin(5*x))
> ##D f21 <- function(x) 0.6*(exp(x) + sin(2.9*x))
> ##D
> ##D ys <- 0.58 + 2.5*bi + f11(x1) + f12(x2) + u[, 1] > 0
> ##D y <- -0.68 - 1.5*bi + f21(x1) + + u[, 2] > 0
> ##D yo <- y*(ys > 0)
> ##D
> ##D dataSim <- data.frame(y, ys, yo, bi, x1, x2)
> ##D
> ##D ## Testing the hypothesis of absence of non-random sample selection...
> ##D
> ##D LM.bpm(list(ys ~ bi + s(x1) + s(x2), yo ~ bi + s(x1)), dataSim, Model = "BSS")
> ##D
> ##D # p-value suggests presence of sample selection, hence fit a bivariate model
> ##D
> ##D #
> ##D ## SEMIPARAMETRIC SAMPLE SELECTION BIVARIATE PROBIT
> ##D ## the first equation MUST be the selection equation
> ##D
> ##D out <- SemiParBIVProbit(list(ys ~ bi + s(x1) + s(x2),
> ##D yo ~ bi + s(x1)),
> ##D data = dataSim, Model = "BSS")
> ##D conv.check(out)
> ##D gt.bpm(out)
> ##D
> ##D ## compare the two summary outputs
> ##D ## the second output produces a summary of the results obtained when
> ##D ## selection bias is not accounted for
> ##D
> ##D summary(out)
> ##D summary(out$gam2)
> ##D
> ##D ## corrected predicted probability that 'yo' is equal to 1
> ##D
> ##D mb(ys, yo, Model = "BSS")
> ##D prev(out, hd.plot = TRUE)
> ##D prev(out, type = "univariate", hd.plot = TRUE)
> ##D
> ##D ## estimated smooth function plots
> ##D ## the red line is the true curve
> ##D ## the blue line is the univariate model curve not accounting for selection bias
> ##D
> ##D x1.s <- sort(x1[dataSim$ys>0])
> ##D f21.x1 <- f21(x1.s)[order(x1.s)]-mean(f21(x1.s))
> ##D
> ##D plot(out, eq = 2, ylim = c(-1.65,0.95)); lines(x1.s, f21.x1, col="red")
> ##D par(new = TRUE)
> ##D plot(out$gam2, se = FALSE, col = "blue", ylim = c(-1.65,0.95),
> ##D ylab = "", rug = FALSE)
> ##D
> ##D #
> ##D #
> ##D ## try a Clayton copula model...
> ##D
> ##D outC <- SemiParBIVProbit(list(ys ~ bi + s(x1) + s(x2),
> ##D yo ~ bi + s(x1)),
> ##D data = dataSim, Model = "BSS", BivD = "C0")
> ##D conv.check(outC)
> ##D summary(outC, cm.plot = TRUE)
> ##D prev(outC)
> ##D
> ##D # in the above examples a third equation could be introduced
> ##D # as illustrated in Example 1
> ##D
> ##D #
> ##D ################
> ##D ## See also ?hiv
> ##D ################
> ##D
> ##D ############
> ##D ## EXAMPLE 4
> ##D ############
> ##D ## Generate data with partial observability
> ##D
> ##D set.seed(0)
> ##D
> ##D n <- 10000
> ##D
> ##D Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
> ##D u <- rMVN(n, rep(0,2), Sigma)
> ##D
> ##D x1 <- round(runif(n)); x2 <- runif(n); x3 <- runif(n)
> ##D
> ##D y1 <- ifelse(-1.55 + 2*x1 + x2 + u[,1] > 0, 1, 0)
> ##D y2 <- ifelse( 0.45 - x3 + u[,2] > 0, 1, 0)
> ##D y <- y1*y2
> ##D
> ##D dataSim <- data.frame(y, x1, x2, x3)
> ##D
> ##D
> ##D ## BIVARIATE PROBIT with Partial Observability
> ##D
> ##D out <- SemiParBIVProbit(list(y ~ x1 + x2,
> ##D y ~ x3),
> ##D data = dataSim, Model = "BPO")
> ##D conv.check(out)
> ##D summary(out)
> ##D
> ##D # first ten estimated probabilities for the four events from object out
> ##D
> ##D cbind(out$p11, out$p10, out$p00, out$p01)[1:10,]
> ##D
> ##D
> ##D # case with smooth function
> ##D # (more computationally intensive)
> ##D
> ##D f1 <- function(x) cos(pi*2*x) + sin(pi*x)
> ##D
> ##D y1 <- ifelse(-1.55 + 2*x1 + f1(x2) + u[,1] > 0, 1, 0)
> ##D y2 <- ifelse( 0.45 - x3 + u[,2] > 0, 1, 0)
> ##D y <- y1*y2
> ##D
> ##D dataSim <- data.frame(y, x1, x2, x3)
> ##D
> ##D out <- SemiParBIVProbit(list(y ~ x1 + s(x2),
> ##D y ~ x3),
> ##D data = dataSim, Model = "BPO")
> ##D
> ##D conv.check(out)
> ##D summary(out, cm.plot = TRUE)
> ##D
> ##D
> ##D # plot estimated and true functions
> ##D
> ##D x2 <- sort(x2); f1.x2 <- f1(x2)[order(x2)] - mean(f1(x2))
> ##D plot(out, eq = 1, scale = 0); lines(x2, f1.x2, col = "red")
> ##D
> ##D #
> ##D ################
> ##D ## See also ?war
> ##D ################
> ##D
> ##D ############
> ##D ## EXAMPLE 5
> ##D ############
> ##D ## Generate data with one endogenous binary variable
> ##D ## and continuous outcome
> ##D
> ##D set.seed(0)
> ##D
> ##D n <- 1000
> ##D
> ##D Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
> ##D u <- rMVN(n, rep(0,2), Sigma)
> ##D
> ##D cov <- rMVN(n, rep(0,2), Sigma)
> ##D cov <- pnorm(cov)
> ##D x1 <- round(cov[,1]); x2 <- cov[,2]
> ##D
> ##D f1 <- function(x) cos(pi*2*x) + sin(pi*x)
> ##D f2 <- function(x) x+exp(-30*(x-0.5)^2)
> ##D
> ##D y1 <- ifelse(-1.55 + 2*x1 + f1(x2) + u[,1] > 0, 1, 0)
> ##D y2 <- -0.25 - 1.25*y1 + f2(x2) + u[,2]
> ##D
> ##D dataSim <- data.frame(y1, y2, x1, x2)
> ##D
> ##D
> ##D ## RECURSIVE Model
> ##D
> ##D rc <- resp.check(y2, margin = "N", print.par = TRUE, loglik = TRUE)
> ##D AIC(rc); BIC(rc)
> ##D
> ##D out <- SemiParBIVProbit(list(y1 ~ x1 + x2,
> ##D y2 ~ y1 + x2),
> ##D data = dataSim, margins = c("probit","N"))
> ##D conv.check(out)
> ##D summary(out)
> ##D post.check(out)
> ##D
> ##D ## SEMIPARAMETRIC RECURSIVE Model
> ##D
> ##D eq.mu.1 <- y1 ~ x1 + s(x2)
> ##D eq.mu.2 <- y2 ~ y1 + s(x2)
> ##D eq.sigma2 <- ~ 1
> ##D eq.theta <- ~ 1
> ##D
> ##D fl <- list(eq.mu.1, eq.mu.2, eq.sigma2, eq.theta)
> ##D
> ##D out <- SemiParBIVProbit(fl, data = dataSim,
> ##D margins = c("probit","N"), gamlssfit = TRUE)
> ##D conv.check(out)
> ##D summary(out)
> ##D post.check(out)
> ##D jc.probs(out, 1, 1.5, intervals = TRUE)[1:4,]
> ##D AT(out, nm.end = "y1")
> ##D AT(out, nm.end = "y1", type = "univariate")
> ##D
> ##D
> ##D #
> ##D #
> ##D
> ##D ############
> ##D ## EXAMPLE 6
> ##D ############
> ##D ## Generate data with one endogenous continuous exposure
> ##D ## and binary outcome
> ##D
> ##D set.seed(0)
> ##D
> ##D n <- 1000
> ##D
> ##D Sigma <- matrix(0.5, 2, 2); diag(Sigma) <- 1
> ##D u <- rMVN(n, rep(0,2), Sigma)
> ##D
> ##D cov <- rMVN(n, rep(0,2), Sigma)
> ##D cov <- pnorm(cov)
> ##D x1 <- round(cov[,1]); x2 <- cov[,2]
> ##D
> ##D f1 <- function(x) cos(pi*2*x) + sin(pi*x)
> ##D f2 <- function(x) x+exp(-30*(x-0.5)^2)
> ##D
> ##D y1 <- -0.25 - 2*x1 + f2(x2) + u[,2]
> ##D y2 <- ifelse(-0.25 - 0.25*y1 + f1(x2) + u[,1] > 0, 1, 0)
> ##D
> ##D dataSim <- data.frame(y1, y2, x1, x2)
> ##D
> ##D eq.mu.1 <- y2 ~ y1 + s(x2)
> ##D eq.mu.2 <- y1 ~ x1 + s(x2)
> ##D eq.sigma2 <- ~ 1
> ##D eq.theta <- ~ 1
> ##D
> ##D fl <- list(eq.mu.1, eq.mu.2, eq.sigma2, eq.theta)
> ##D
> ##D out <- SemiParBIVProbit(fl, data = dataSim,
> ##D margins = c("probit","N"))
> ##D conv.check(out)
> ##D summary(out)
> ##D post.check(out)
> ##D AT(out, nm.end = "y1")
> ##D AT(out, nm.end = "y1", type = "univariate")
> ##D RR(out, nm.end = "y1", rr.plot = TRUE)
> ##D RR(out, nm.end = "y1", type = "univariate")
> ##D OR(out, nm.end = "y1", or.plot = TRUE)
> ##D OR(out, nm.end = "y1", type = "univariate")
> ##D
> ## End(Not run)
>
>
>
>
>
> dev.off()
null device
1
>
|