Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
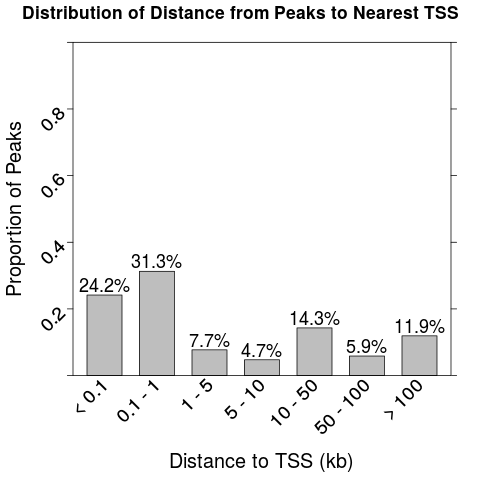
Plot histogram of distance from peak to nearest TSSDescriptionCreate a histogram of the distance from each peak to the nearest transcription start site (TSS) of any gene. Usageplot_dist_to_tss(peaks, genome = "hg19") Arguments
ValueA trellis plot object. Author(s)Ryan Welch welchr@umich.edu See Also
Exampleslibrary(chipenrich.data) library(chipenrich) # Create histogram of distance from peaks to nearest TSS. data(peaks_E2F4) plot_dist_to_tss(peaks_E2F4) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(chipenrich)
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/chipenrich/plot_dist_to_tss.Rd_%03d_medium.png", width=480, height=480)
> ### Name: plot_dist_to_tss
> ### Title: Plot histogram of distance from peak to nearest TSS
> ### Aliases: plot_dist_to_tss
>
> ### ** Examples
>
> library(chipenrich.data)
> library(chipenrich)
>
> # Create histogram of distance from peaks to nearest TSS.
> data(peaks_E2F4)
> plot_dist_to_tss(peaks_E2F4)
>
>
>
>
>
> dev.off()
null device
1
>
|