Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
1-df and 2-df tests for genetic associations with SNPsDescriptionThis function carries out tests for association between phenotype and a series of single nucleotide polymorphisms (SNPs), within strata defined by a possibly confounding factor. SNPs are considered one at a time and both 1-df and 2-df tests are calculated. For a binary phenotype, the 1-df test is the Cochran-Armitage test (or, when stratified, the Mantel-extension test). Usagesingle.snp.tests(phenotype, stratum, data = sys.parent(), snp.data, subset, snp.subset) Arguments
DetailsFormally, the test statistics are score tests for generalized linear models with canonical link. That is, they are inner products between genotype indicators and the deviations of phenotypes from their stratum means. Variances (and covariances) are those of the permutation distribution obtained by randomly permuting phenotype within stratum. The ValueA dataframe, with columns
NoteThe behaviour of this function for objects of class
Author(s)David Clayton david.clayton@cimr.cam.ac.uk ReferencesClayton (2008) Testing for association on the X chromosome Biostatistics (In press) See Also
Examples
data(testdata)
results <- single.snp.tests(cc, stratum=region, data=subject.data,
snp.data=Autosomes, snp.subset=1:10)
summary(results)
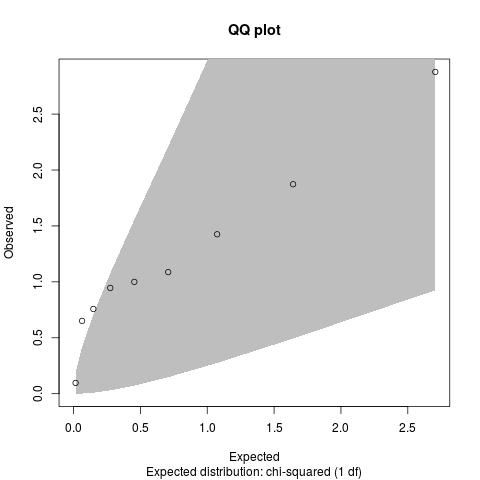
# QQ plot - see code{link{qq.chisq}}
qq.chisq(results$chi2.1df)
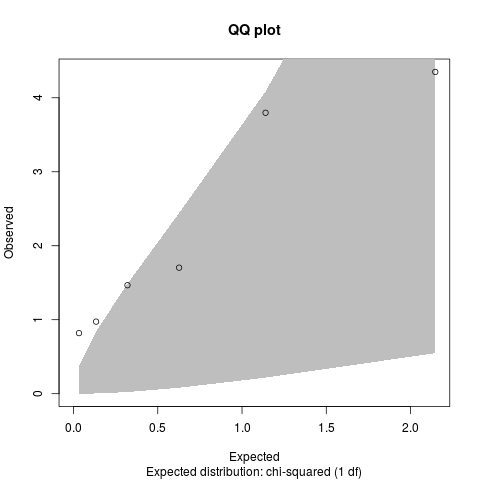
qq.chisq(results$chi2.2df)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(chopsticks)
Loading required package: survival
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/chopsticks/single.snp.tests.Rd_%03d_medium.png", width=480, height=480)
> ### Name: single.snp.tests
> ### Title: 1-df and 2-df tests for genetic associations with SNPs
> ### Aliases: single.snp.tests
> ### Keywords: htest
>
> ### ** Examples
>
> data(testdata)
> results <- single.snp.tests(cc, stratum=region, data=subject.data,
+ snp.data=Autosomes, snp.subset=1:10)
> summary(results)
chi2.1df chi2.2df p.1df p.2df
Min. :0.0963 Min. :0.8189 Min. :0.08984 Min. :0.1136
1st Qu.:0.7577 1st Qu.:1.0972 1st Qu.:0.23253 1st Qu.:0.2190
Median :1.0000 Median :1.5847 Median :0.31731 Median :0.4536
Mean :1.1903 Mean :2.1849 Mean :0.33322 Mean :0.4082
3rd Qu.:1.4253 3rd Qu.:3.2740 3rd Qu.:0.38406 3rd Qu.:0.5809
Max. :2.8773 Max. :4.3497 Max. :0.75631 Max. :0.6640
NA's :1 NA's :4 NA's :1 NA's :4
N
Min. :378.0
1st Qu.:396.2
Median :398.5
Mean :395.6
3rd Qu.:400.0
Max. :400.0
> # QQ plot - see code{qq.chisq}
> qq.chisq(results$chi2.1df)
N omitted lambda
9.000000 0.000000 4.867221
> qq.chisq(results$chi2.2df)
N omitted lambda
6.00000 0.00000 6.69583
>
>
>
>
>
> dev.off()
null device
1
>
|