Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Comparison of two flat clusteringsDescription
Usage
flatVSflat(weights, coord1 = NULL, coord2 = NULL, max.iter = 24, h.min = 0.1,
plotting = TRUE, horiz = FALSE, offset = 0.1, line.wd = 3, point.sz = 2,
evenly = FALSE, main = "", xlab = "", ylab = "", col = NULL, ...)
Arguments
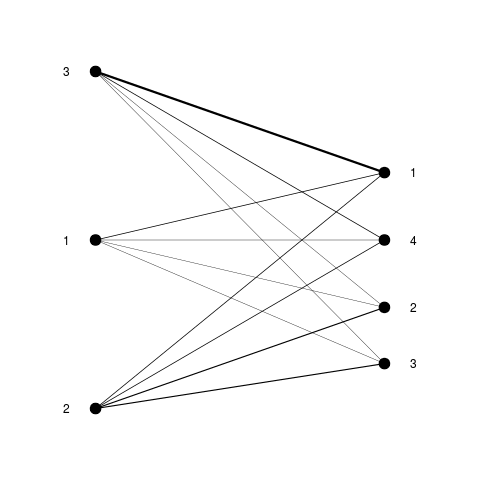
DetailsAs the iterations of the algorithm run the coordinates of the nodes in a
single layer are updated. For a given partition, each node is assigned a
new position, the gravity-centre, using the barycentre algorithm; then, the
nodes in the corresponding layer are reordered according to the new
positions. If the gravity-centres cause two consecutive nodes to be less
than Valuea list of components including:
Author(s)Aurora Torrente aurora@ebi.ac.uk and Alvis Brazma brazma@ebi.ac.uk ReferencesEades, P. et al. (1986). On an edge crossing problem. Proc. of 9th Australian Computer Science Conference, pp. 327-334. Gansner, E.R. et al. (1993). A technique for drawing directed graphs. IEEE Trans. on Software Engineering, 19 (3), 214-230. Garey, M.R. et al. (1983). Crossing number in NP complete. SIAM J. Algebraic Discrete Methods, 4, 312-316. Torrente, A. et al. (2005). A new algorithm for comparing and visualizing relationships between hierarchical and flat gene expression data clusterings. Bioinformatics, 21 (21), 3993-3999. See AlsoflatVShier, barycentre Examples
# simulated data
clustering1 <- c(rep(1, 5), rep(2, 10), rep(3, 10))
clustering2 <- c(rep(1:4, 5), rep(1, 5))
weights <- table(clustering1, clustering2)
flatVSflat(table(clustering1, clustering2))
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(clustComp)
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/clustComp/flatVSflat.Rd_%03d_medium.png", width=480, height=480)
> ### Name: flatVSflat
> ### Title: Comparison of two flat clusterings
> ### Aliases: flatVSflat
> ### Keywords: clustering comparison
>
> ### ** Examples
>
> # simulated data
> clustering1 <- c(rep(1, 5), rep(2, 10), rep(3, 10))
> clustering2 <- c(rep(1:4, 5), rep(1, 5))
> weights <- table(clustering1, clustering2)
> flatVSflat(table(clustering1, clustering2))
$icross
[1] 91
$fcross
[1] 39
$coord1
1 2 3
0.8 0.5 1.1
$coord2
1 2 3 4
0.92 0.68 0.58 0.80
>
>
>
>
>
> dev.off()
null device
1
>
|