Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Creating Filters and Filtering Flow Cytometry DataDescriptionThe UsagetmixFilter(filterId="tmixFilter", parameters="", ...) Arguments
ValueThe The NoteThe If If Author(s)Raphael Gottardo <raph@stat.ubc.ca>, Kenneth Lo <c.lo@stat.ubc.ca> ReferencesLo, K., Brinkman, R. R. and Gottardo, R. (2008) Automated Gating of Flow Cytometry Data via Robust Model-based Clustering. Cytometry A 73, 321-332. See Also
Examples
### The example below largely resembles the one in the flowClust
### man page. The main purpose here is to demonstrate how the
### entire cluster analysis can be done in a fashion highly
### integrated into flowCore.
data(rituximab)
### create a filter object
s1filter <- tmixFilter("s1", c("FSC.H", "SSC.H"), K=1)
### cluster the data using FSC.H and SSC.H
res1 <- filter(rituximab, s1filter)
### remove outliers before proceeding to the second stage
# %in% operator returns a logical vector indicating whether each
# of the observations lies inside the gate or not
rituximab2 <- rituximab[rituximab %in% res1,]
# a shorthand for the above line
rituximab2 <- rituximab[res1,]
# this can also be done using the Subset method
rituximab2 <- Subset(rituximab, res1)
### cluster the data using FL1.H and FL3.H (with 3 clusters)
s2filter <- tmixFilter("s2", c("FL1.H", "FL3.H"), K=3)
res2 <- filter(rituximab2, s2filter)
show(s2filter)
show(res2)
summary(res2)
# to demonstrate the use of the split method
split(rituximab2, res2)
split(rituximab2, res2, population=list(sc1=c(1,2), sc2=3))
# to show the cluster assignment of observations
table(Map(res2))
# to show the cluster centres (i.e., the mean parameter estimates
# transformed back to the original scale) and proportions
getEstimates(res2)
### demonstrate the use of various plotting methods
# a scatterplot
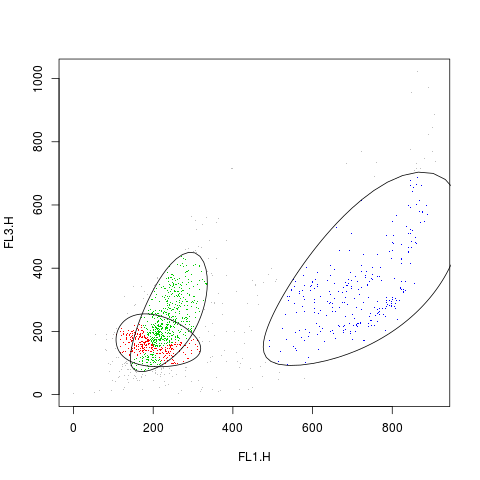
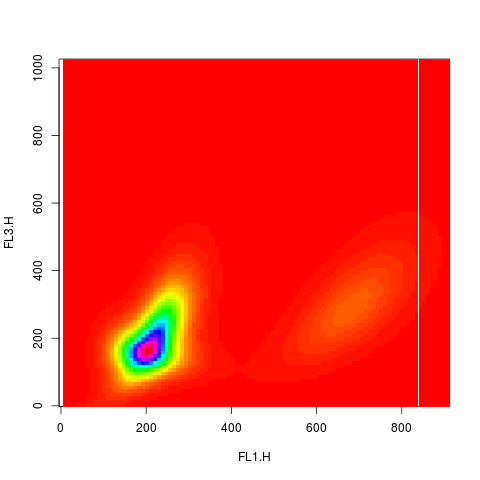
plot(rituximab2, res2, level=0.8)
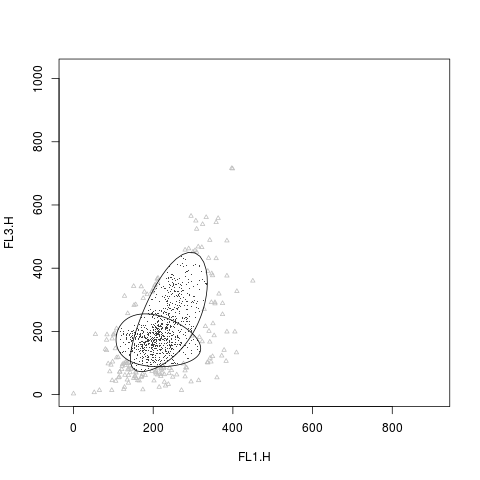
plot(rituximab2, res2, level=0.8, include=c(1,2), grayscale=TRUE,
pch.outliers=2)
# a contour / image plot
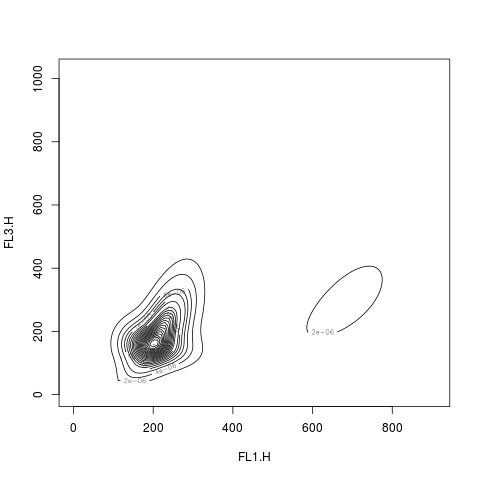
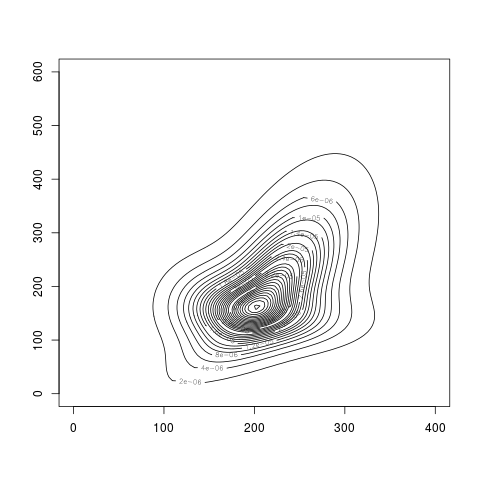
res2.den <- density(res2, data=rituximab2)
plot(res2.den)
plot(res2.den, scale="sqrt", drawlabels=FALSE)
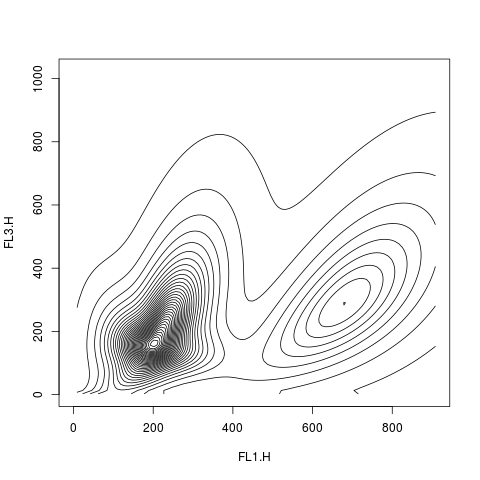
plot(res2.den, type="image", nlevels=100)
plot(density(res2, include=c(1,2), from=c(0,0), to=c(400,600)))
# a histogram (1-D density) plot
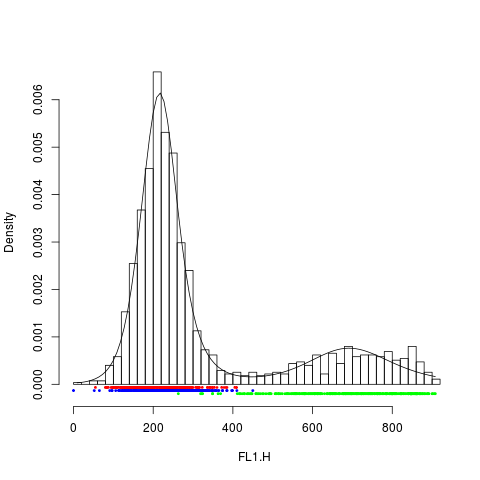
plot(rituximab2, res2, "FL1.H")
### to demonstrate the use of the ruleOutliers method
summary(res2)
# change the rule to call outliers
ruleOutliers(res2) <- list(level=0.95)
# augmented cluster boundaries lead to fewer outliers
summary(res2)
# the following line illustrates how to select a subset of data
# to perform cluster analysis through the min and max arguments;
# also note the use of level to specify a rule to call outliers
# other than the default
s2t <- tmixFilter("s2t", c("FL1.H", "FL3.H"), K=3, B=100,
min=c(0,0), max=c(400,800), level=0.95, z.cutoff=0.5)
filter(rituximab2, s2t)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(flowClust)
Loading required package: Biobase
Loading required package: BiocGenerics
Loading required package: parallel
Attaching package: 'BiocGenerics'
The following objects are masked from 'package:parallel':
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLB
The following objects are masked from 'package:stats':
IQR, mad, xtabs
The following objects are masked from 'package:base':
Filter, Find, Map, Position, Reduce, anyDuplicated, append,
as.data.frame, cbind, colnames, do.call, duplicated, eval, evalq,
get, grep, grepl, intersect, is.unsorted, lapply, lengths, mapply,
match, mget, order, paste, pmax, pmax.int, pmin, pmin.int, rank,
rbind, rownames, sapply, setdiff, sort, table, tapply, union,
unique, unsplit
Welcome to Bioconductor
Vignettes contain introductory material; view with
'browseVignettes()'. To cite Bioconductor, see
'citation("Biobase")', and for packages 'citation("pkgname")'.
Loading required package: graph
Loading required package: RBGL
Loading required package: ellipse
Loading required package: flowViz
Loading required package: flowCore
Attaching package: 'flowCore'
The following object is masked from 'package:BiocGenerics':
normalize
Loading required package: lattice
Loading required package: mnormt
Loading required package: corpcor
Loading required package: clue
Attaching package: 'flowClust'
The following object is masked from 'package:graphics':
box
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/flowClust/tmixFilter.Rd_%03d_medium.png", width=480, height=480)
> ### Name: tmixFilter
> ### Title: Creating Filters and Filtering Flow Cytometry Data
> ### Aliases: tmixFilter filter,flowFrame,tmixFilter-method
> ### filter,flowFrame,filter-method filter,flowFrame,filterSet-method
> ### filter,flowSet,filter-method filter,flowSet,filterSet-method
> ### filter,flowSet,list-method filter-method filter.flowFrame filter
> ### tmixFilter-class tmixFilterResult-class tmixFilterResultList
> ### tmixFilterResultList-class %in%,flowFrame,tmixFilter-method
> ### %in%,ANY,tmixFilterResult-method
> ### %in%,flowFrame,tmixFilterResult-method
> ### %in%,ANY,tmixFilterResultList-method
> ### [,flowFrame,tmixFilterResult-method
> ### [,flowFrame,tmixFilterResultList-method
> ### [[,tmixFilterResultList,ANY-method
> ### [,flowFrame,tmixFilterResult,ANY,ANY-method
> ### [,flowFrame,tmixFilterResultList,ANY,ANY-method
> ### Keywords: cluster models
>
> ### ** Examples
>
> ### The example below largely resembles the one in the flowClust
> ### man page. The main purpose here is to demonstrate how the
> ### entire cluster analysis can be done in a fashion highly
> ### integrated into flowCore.
>
>
> data(rituximab)
>
> ### create a filter object
> s1filter <- tmixFilter("s1", c("FSC.H", "SSC.H"), K=1)
> ### cluster the data using FSC.H and SSC.H
> res1 <- filter(rituximab, s1filter)
The prior specification has no effect when usePrior=no
Using the parallel (multicore) version of flowClust with 4 cores
>
> ### remove outliers before proceeding to the second stage
> # %in% operator returns a logical vector indicating whether each
> # of the observations lies inside the gate or not
> rituximab2 <- rituximab[rituximab %in% res1,]
> # a shorthand for the above line
> rituximab2 <- rituximab[res1,]
> # this can also be done using the Subset method
> rituximab2 <- Subset(rituximab, res1)
>
> ### cluster the data using FL1.H and FL3.H (with 3 clusters)
> s2filter <- tmixFilter("s2", c("FL1.H", "FL3.H"), K=3)
> res2 <- filter(rituximab2, s2filter)
The prior specification has no effect when usePrior=no
Using the parallel (multicore) version of flowClust with 4 cores
>
> show(s2filter)
A t-mixture filter named 's2': FL1.H, FL3.H
** Clustering Settings **
Number of clusters: 3
Degrees of freedom of t distribution: 4
Maximum number of EM iterations: 500
Transformation selection will be performed
Rule of identifying outliers: 90% quantile
> show(res2)
Object of class 'tmixFilterResult'
This object stores the result of applying a t-mixture filternamed 's2' on A02, which has an experiment name 'Flow Experiment'
The following parameters have been used in filtering:
FL1.H, FL3.H
The 'tmixFilterResult' class extends the 'flowClust' class, which contains the following slots:
expName, varNames, K, w, mu, sigma, lambda, nu, z, u, label, uncertainty, ruleOutliers, flagOutliers, rm.min, rm.max, logLike, BIC, ICL
> summary(res2)
** Experiment Information **
Experiment name: Flow Experiment
Variables used: FL1.H FL3.H
** Clustering Summary **
Number of clusters: 3
Proportions: 0.2658702 0.5091045 0.2250253
** Transformation Parameter **
lambda: 0.4312673
** Information Criteria **
Log likelihood: -16475.41
BIC: -33080.88
ICL: -34180.67
** Data Quality **
Number of points filtered from above: 0 (0%)
Number of points filtered from below: 0 (0%)
Rule of identifying outliers: 90% quantile
Number of outliers: 96 (6.99%)
Uncertainty summary:
>
> # to demonstrate the use of the split method
> split(rituximab2, res2)
[[1]]
flowFrame object 'A02 (1)'
with 320 cells and 8 observables:
name desc range maxRange minRange
$P1 FSC.H FSC-Height 1024 1023 0
$P2 SSC.H Side Scatter 1024 1023 0
$P3 FL1.H Anti-BrdU FITC 1024 1023 0
$P4 FL2.H <NA> 1024 1023 0
$P5 FL3.H 7 AAD 1024 1023 0
$P6 FL1.A <NA> 1024 1023 0
$P7 FL1.W <NA> 1024 1023 0
$P8 Time Time (204.80 sec.) 1024 1023 0
136 keywords are stored in the 'description' slot
[[2]]
flowFrame object 'A02 (2)'
with 670 cells and 8 observables:
name desc range maxRange minRange
$P1 FSC.H FSC-Height 1024 1023 0
$P2 SSC.H Side Scatter 1024 1023 0
$P3 FL1.H Anti-BrdU FITC 1024 1023 0
$P4 FL2.H <NA> 1024 1023 0
$P5 FL3.H 7 AAD 1024 1023 0
$P6 FL1.A <NA> 1024 1023 0
$P7 FL1.W <NA> 1024 1023 0
$P8 Time Time (204.80 sec.) 1024 1023 0
136 keywords are stored in the 'description' slot
[[3]]
flowFrame object 'A02 (3)'
with 288 cells and 8 observables:
name desc range maxRange minRange
$P1 FSC.H FSC-Height 1024 1023 0
$P2 SSC.H Side Scatter 1024 1023 0
$P3 FL1.H Anti-BrdU FITC 1024 1023 0
$P4 FL2.H <NA> 1024 1023 0
$P5 FL3.H 7 AAD 1024 1023 0
$P6 FL1.A <NA> 1024 1023 0
$P7 FL1.W <NA> 1024 1023 0
$P8 Time Time (204.80 sec.) 1024 1023 0
136 keywords are stored in the 'description' slot
> split(rituximab2, res2, population=list(sc1=c(1,2), sc2=3))
$sc1
flowFrame object 'A02 (1,2)'
with 990 cells and 8 observables:
name desc range maxRange minRange
$P1 FSC.H FSC-Height 1024 1023 0
$P2 SSC.H Side Scatter 1024 1023 0
$P3 FL1.H Anti-BrdU FITC 1024 1023 0
$P4 FL2.H <NA> 1024 1023 0
$P5 FL3.H 7 AAD 1024 1023 0
$P6 FL1.A <NA> 1024 1023 0
$P7 FL1.W <NA> 1024 1023 0
$P8 Time Time (204.80 sec.) 1024 1023 0
3 keywords are stored in the 'description' slot
$sc2
flowFrame object 'A02 (3)'
with 288 cells and 8 observables:
name desc range maxRange minRange
$P1 FSC.H FSC-Height 1024 1023 0
$P2 SSC.H Side Scatter 1024 1023 0
$P3 FL1.H Anti-BrdU FITC 1024 1023 0
$P4 FL2.H <NA> 1024 1023 0
$P5 FL3.H 7 AAD 1024 1023 0
$P6 FL1.A <NA> 1024 1023 0
$P7 FL1.W <NA> 1024 1023 0
$P8 Time Time (204.80 sec.) 1024 1023 0
136 keywords are stored in the 'description' slot
>
> # to show the cluster assignment of observations
> table(Map(res2))
1 2 3
320 670 288
>
> # to show the cluster centres (i.e., the mean parameter estimates
> # transformed back to the original scale) and proportions
> getEstimates(res2)
$proportions
[1] 0.2658702 0.5091045 0.2250253
$locations
[,1] [,2]
[1,] 196.7897 159.5326
[2,] 227.7338 215.6581
[3,] 699.9997 316.2946
>
> ### demonstrate the use of various plotting methods
> # a scatterplot
> plot(rituximab2, res2, level=0.8)
Rule of identifying outliers: 80% quantile
> plot(rituximab2, res2, level=0.8, include=c(1,2), grayscale=TRUE,
+ pch.outliers=2)
Rule of identifying outliers: 80% quantile
> # a contour / image plot
> res2.den <- density(res2, data=rituximab2)
> plot(res2.den)
> plot(res2.den, scale="sqrt", drawlabels=FALSE)
> plot(res2.den, type="image", nlevels=100)
> plot(density(res2, include=c(1,2), from=c(0,0), to=c(400,600)))
> # a histogram (1-D density) plot
> plot(rituximab2, res2, "FL1.H")
>
> ### to demonstrate the use of the ruleOutliers method
> summary(res2)
** Experiment Information **
Experiment name: Flow Experiment
Variables used: FL1.H FL3.H
** Clustering Summary **
Number of clusters: 3
Proportions: 0.2658702 0.5091045 0.2250253
** Transformation Parameter **
lambda: 0.4312673
** Information Criteria **
Log likelihood: -16475.41
BIC: -33080.88
ICL: -34180.67
** Data Quality **
Number of points filtered from above: 0 (0%)
Number of points filtered from below: 0 (0%)
Rule of identifying outliers: 90% quantile
Number of outliers: 96 (6.99%)
Uncertainty summary:
> # change the rule to call outliers
> ruleOutliers(res2) <- list(level=0.95)
Rule of identifying outliers: 95% quantile
> # augmented cluster boundaries lead to fewer outliers
> summary(res2)
** Experiment Information **
Experiment name: Flow Experiment
Variables used: FL1.H FL3.H
** Clustering Summary **
Number of clusters: 3
Proportions: 0.2658702 0.5091045 0.2250253
** Transformation Parameter **
lambda: 0.4312673
** Information Criteria **
Log likelihood: -16475.41
BIC: -33080.88
ICL: -34180.67
** Data Quality **
Number of points filtered from above: 0 (0%)
Number of points filtered from below: 0 (0%)
Rule of identifying outliers: 95% quantile
Number of outliers: 35 (2.55%)
Uncertainty summary:
>
> # the following line illustrates how to select a subset of data
> # to perform cluster analysis through the min and max arguments;
> # also note the use of level to specify a rule to call outliers
> # other than the default
> s2t <- tmixFilter("s2t", c("FL1.H", "FL3.H"), K=3, B=100,
+ min=c(0,0), max=c(400,800), level=0.95, z.cutoff=0.5)
> filter(rituximab2, s2t)
The prior specification has no effect when usePrior=no
Using the parallel (multicore) version of flowClust with 4 cores
Object of class 'tmixFilterResult'
This object stores the result of applying a t-mixture filternamed 's2t' on A02, which has an experiment name 'Flow Experiment'
The following parameters have been used in filtering:
FL1.H, FL3.H
The 'tmixFilterResult' class extends the 'flowClust' class, which contains the following slots:
expName, varNames, K, w, mu, sigma, lambda, nu, z, u, label, uncertainty, ruleOutliers, flagOutliers, rm.min, rm.max, logLike, BIC, ICL
>
>
>
>
>
> dev.off()
null device
1
>
|