|
Last data update: 2014.03.03
|
R: critical regions
| critVal.target | R Documentation |
critical regions
Description
critical region cutpoints
Usage
critVal.target(k, p0, target, posdiff = NULL, ns)
Arguments
k |
window width(s)
|
p0 |
length 2 probabilities
|
target |
- two tailed
|
posdiff |
- position difference matrix
|
ns |
the number of windows passing filter at each k
|
Details
This version uses TFD and will find alpha implicitly
Value
list of cutoffs and attributes
Author(s)
Charles Berry
See Also
gRxCluster for how and why this function is
used
Examples
# symmetric odds:
crit <- critVal.target(5:25,c(1,1),1,ns=rep(10,21))
crit[[1]]
sapply(crit,c)
# 5:1 odds
asymmetric.crit <- critVal.target(5:25,c(1,5),1,ns=rep(10,21))
# show the critical regions
par(mfrow=c(1,2))
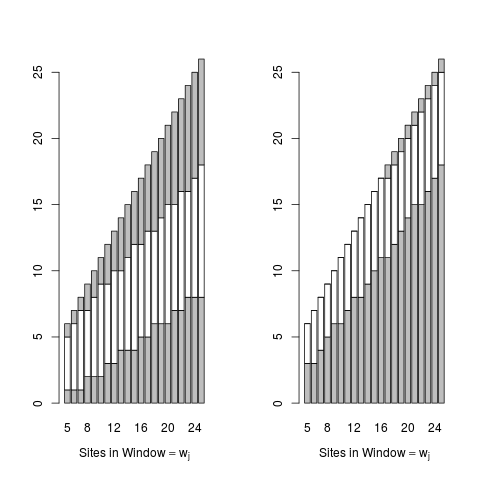
gRxPlot(crit,method="critical")
gRxPlot(asymmetric.crit,method="critical")
rm(crit,asymmetric.crit)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(geneRxCluster)
Loading required package: GenomicRanges
Loading required package: BiocGenerics
Loading required package: parallel
Attaching package: 'BiocGenerics'
The following objects are masked from 'package:parallel':
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLB
The following objects are masked from 'package:stats':
IQR, mad, xtabs
The following objects are masked from 'package:base':
Filter, Find, Map, Position, Reduce, anyDuplicated, append,
as.data.frame, cbind, colnames, do.call, duplicated, eval, evalq,
get, grep, grepl, intersect, is.unsorted, lapply, lengths, mapply,
match, mget, order, paste, pmax, pmax.int, pmin, pmin.int, rank,
rbind, rownames, sapply, setdiff, sort, table, tapply, union,
unique, unsplit
Loading required package: S4Vectors
Loading required package: stats4
Attaching package: 'S4Vectors'
The following objects are masked from 'package:base':
colMeans, colSums, expand.grid, rowMeans, rowSums
Loading required package: IRanges
Loading required package: GenomeInfoDb
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/geneRxCluster/critVal.target.Rd_%03d_medium.png", width=480, height=480)
> ### Name: critVal.target
> ### Title: critical regions
> ### Aliases: critVal.target
>
> ### ** Examples
>
> # symmetric odds:
> crit <- critVal.target(5:25,c(1,1),1,ns=rep(10,21))
> crit[[1]]
low up
1 5
attr(,"fdr")
target.low target.hi low hi
[1,] 0.625 20.000 0.03125 1.00000
[2,] 3.750 19.375 0.18750 0.96875
[3,] 10.000 16.250 0.50000 0.81250
[4,] 16.250 10.000 0.81250 0.50000
[5,] 19.375 3.750 0.96875 0.18750
[6,] 20.000 0.625 1.00000 0.03125
attr(,"target")
[1] 1
> sapply(crit,c)
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14]
low 1 1 1 2 2 2 3 3 4 4 4 5 5 6
up 5 6 7 7 8 9 9 10 10 11 12 12 13 13
[,15] [,16] [,17] [,18] [,19] [,20] [,21]
low 6 6 7 7 8 8 8
up 14 15 15 16 16 17 18
> # 5:1 odds
> asymmetric.crit <- critVal.target(5:25,c(1,5),1,ns=rep(10,21))
> # show the critical regions
> par(mfrow=c(1,2))
> gRxPlot(crit,method="critical")
> gRxPlot(asymmetric.crit,method="critical")
> rm(crit,asymmetric.crit)
>
>
>
>
>
> dev.off()
null device
1
>
|
|
 providing
providing  .
.