Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Estimate and print the copy number profile of some chromosomes of a sampleDescriptionFunction to estimate the copy number profile with a piecewise constant function using mBPCR. Eventually, it is possible to estimate the profile with a
smoothing curve, using either the Bayesian Regression Curve with K_2 (BRC with K_2) or the Bayesian Regression Curve Averaging over k (BRCAk). It is also possible
to choose the estimator of the variance of the levels Usage
estProfileWithMBPCR(snpName, chr, position, logratio, chrToBeAnalyzed, maxProbeNumber,
rhoSquare=NULL, kMax=50, nu=NULL, sigmaSquare=NULL, typeEstRho=1, regr=NULL)
Arguments
DetailsBy default, the function estimates the copy number profile with mBPCR and estimating rhoSquare on the sample, using hat{ρ}_1^2. It is
also possible to use hat{ρ}^2 as estimator of The function gives also the possibility to estimate the profile with a Bayesian regression curve: if See function ValueA list containing:
ReferencesRancoita, P. M. V., Hutter, M., Bertoni, F., Kwee, I. (2009). Bayesian DNA copy number analysis. BMC Bioinformatics 10: 10. http://www.idsia.ch/~paola/mBPCR See Also
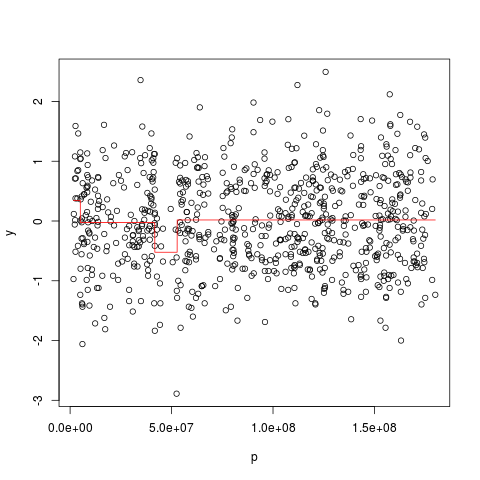
Examples##import the 10K data of cell line REC data(rec10k) ##estimation of the profile of chromosome 5 results <- estProfileWithMBPCR(rec10k$SNPname, rec10k$Chromosome, rec10k$PhysicalPosition, rec10k$log2ratio, chrToBeAnalyzed=5, maxProbeNumber=2000) ##plot the estimated profile of chromosome 5 y <- rec10k$log2ratio[rec10k$Chromosome == 5] p <- rec10k$PhysicalPosition[rec10k$Chromosome == 5] plot(p, y) points(p, results$estPC[rec10k$Chromosome == 5], type='l', col='red') ###for the estimation of the profile of all chromosomes #results <- estProfileWithMBPCR(rec10k$SNPname, rec10k$Chromosome, rec10k$PhysicalPosition, rec10k$log2ratio, chrToBeAnalyzed=c(1:22,'X'), maxProbeNumber=2000) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(mBPCR)
Loading required package: oligoClasses
Welcome to oligoClasses version 1.34.0
Loading required package: SNPchip
Welcome to SNPchip version 2.18.0
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/mBPCR/estProfileWithMBPCR.Rd_%03d_medium.png", width=480, height=480)
> ### Name: estProfileWithMBPCR
> ### Title: Estimate and print the copy number profile of some chromosomes
> ### of a sample
> ### Aliases: estProfileWithMBPCR
> ### Keywords: regression smooth
>
> ### ** Examples
>
> ##import the 10K data of cell line REC
> data(rec10k)
> ##estimation of the profile of chromosome 5
> results <- estProfileWithMBPCR(rec10k$SNPname, rec10k$Chromosome, rec10k$PhysicalPosition, rec10k$log2ratio, chrToBeAnalyzed=5, maxProbeNumber=2000)
Estimation of global parameters
Estimation of the profile of chromosome 5
Computation of log(A^0)
Computation of left and right recursions
Determination of PC Regression
> ##plot the estimated profile of chromosome 5
> y <- rec10k$log2ratio[rec10k$Chromosome == 5]
> p <- rec10k$PhysicalPosition[rec10k$Chromosome == 5]
> plot(p, y)
> points(p, results$estPC[rec10k$Chromosome == 5], type='l', col='red')
>
> ###for the estimation of the profile of all chromosomes
> #results <- estProfileWithMBPCR(rec10k$SNPname, rec10k$Chromosome, rec10k$PhysicalPosition, rec10k$log2ratio, chrToBeAnalyzed=c(1:22,'X'), maxProbeNumber=2000)
>
>
>
>
>
>
> dev.off()
null device
1
>
|