Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Find optimal single feature classifiersDescriptionRun the Messina algorithm to find features (eg. genes) that optimally distinguish between two classes of samples, subject to minimum performance requirements. Usagemessina(x, y, min_sens, min_spec, f_train = 0.9, n_boot = 50, seed = NULL, progress = TRUE, silent = FALSE) Arguments
DetailsNote: If you wish to use Messina to detect differential
expression, and not construct classifiers, you may find the
Messina constructs single-feature threshold classifiers (see below) to separate two sample groups, that are in a sense the most robust single-gene classifiers that satisfy user-supplied performance requirements. It accepts as primary input a matrix or ExpressionSet of feature data x; a vector of sample class membership y; and minimum classifier target performance values min_sens, and min_spec. Messina then examines each feature of x in turn, and attempts to build a threshold classifier that satisfies the minimum performance requirements, based on that feature. The results of this classifier training and testing are then returned in a MessinaClassResult object. The features measured in x must be numeric and contain no missing values, but apart from that are unrestricted – common use cases are mRNA measurements and protein abundance estimates. Messina is not sensitive to the data transformation used, although for mRNA abundance measurements a log-transform or similar is suggested to aid interpretability of the results. x containing discrete values can also be examined by Messina, though if the number of possible values of the members of x is very low, the algorithm is unlikely to be very powerful. Valuean object of class "MessinaClassResult" containing the results of the analysis. Threshold classifiersMessina trains single-feature threshold classifiers. These are classifiers that place unknown samples into one of two groups, based on whether the sample's measurement for a given feature is above or below a constant threshold value. They are the one-dimensional version of support vector machines (SVMs), where in this case the feature set is one-dimensional, and the 'support vector' (the threshold) is a zero-dimensional point. Threshold classifiers are defined by two properties: their threshold value, and their direction, which is the class assigned if a sample's measurement exceeds the threshold. Author(s)Mark Pinese m.pinese@garvan.org.au ReferencesPinese M, Scarlett CJ, Kench JG, et al. (2009) Messina: A Novel Analysis Tool to Identify Biologically Relevant Molecules in Disease. PLoS ONE 4(4): e5337. doi:10.1371/journal.pone.0005337 See Also
Examples
## Load some example data
library(antiProfilesData)
data(apColonData)
x = exprs(apColonData)
y = pData(apColonData)$SubType
## Subset the data to only tumour and normal samples
sel = y %in% c("normal", "tumor")
x = x[,sel]
y = y[sel]
## Run Messina to rank probesets on their classification ability, with
## classifiers needing to meet a minimum sensitivity of 0.95, and minimum
## specificity of 0.85.
fit = messina(x, y == "tumor", min_sens = 0.95, min_spec = 0.85)
## Display the results.
fit
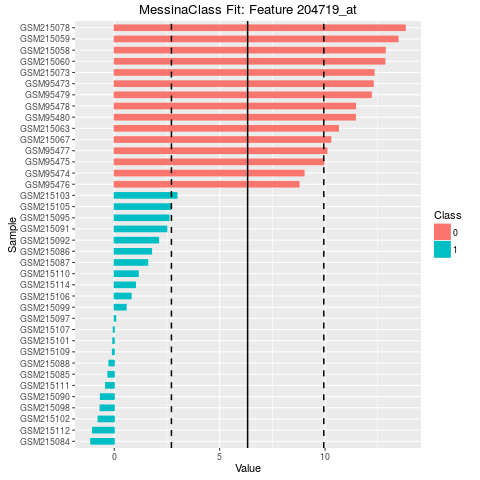
plot(fit)
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(messina)
Loading required package: survival
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/messina/messina.Rd_%03d_medium.png", width=480, height=480)
> ### Name: messina
> ### Title: Find optimal single feature classifiers
> ### Aliases: messina
>
> ### ** Examples
>
> ## Load some example data
> library(antiProfilesData)
Loading required package: Biobase
Loading required package: BiocGenerics
Loading required package: parallel
Attaching package: 'BiocGenerics'
The following objects are masked from 'package:parallel':
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLB
The following objects are masked from 'package:stats':
IQR, mad, xtabs
The following objects are masked from 'package:base':
Filter, Find, Map, Position, Reduce, anyDuplicated, append,
as.data.frame, cbind, colnames, do.call, duplicated, eval, evalq,
get, grep, grepl, intersect, is.unsorted, lapply, lengths, mapply,
match, mget, order, paste, pmax, pmax.int, pmin, pmin.int, rank,
rbind, rownames, sapply, setdiff, sort, table, tapply, union,
unique, unsplit
Welcome to Bioconductor
Vignettes contain introductory material; view with
'browseVignettes()'. To cite Bioconductor, see
'citation("Biobase")', and for packages 'citation("pkgname")'.
> data(apColonData)
>
> x = exprs(apColonData)
> y = pData(apColonData)$SubType
>
> ## Subset the data to only tumour and normal samples
> sel = y %in% c("normal", "tumor")
> x = x[,sel]
> y = y[sel]
>
> ## Run Messina to rank probesets on their classification ability, with
> ## classifiers needing to meet a minimum sensitivity of 0.95, and minimum
> ## specificity of 0.85.
> fit = messina(x, y == "tumor", min_sens = 0.95, min_spec = 0.85)
Performance bootstrapping...
0% [ ] 2% [= ] 4% [== ] 6% [=== ] 8% [==== ] 9% [===== ] 11% [====== ] 13% [======= ] 15% [========= ] 17% [========== ] 19% [=========== ] 21% [============ ] 22% [============= ] 24% [============== ] 26% [=============== ] 28% [================ ] 30% [================= ] 32% [=================== ] 34% [==================== ] 36% [===================== ] 37% [====================== ] 39% [======================= ] 41% [======================== ] 43% [========================= ] 45% [========================== ] 47% [============================ ] 49% [============================= ] 51% [============================== ] 52% [=============================== ] 54% [================================ ] 56% [================================= ] 58% [================================== ] 60% [=================================== ] 62% [===================================== ] 64% [====================================== ] 66% [======================================= ] 67% [======================================== ] 69% [========================================= ] 71% [========================================== ] 73% [=========================================== ] 75% [============================================ ] 77% [============================================== ] 79% [=============================================== ] 81% [================================================ ] 82% [================================================= ] 84% [================================================== ] 86% [=================================================== ] 88% [==================================================== ] 90% [===================================================== ] 92% [======================================================= ] 94% [======================================================== ] 96% [========================================================= ] 97% [========================================================== ] 99% [=========================================================== ] 100% [============================================================]
>
> ## Display the results.
> fit
An object of class MessinaClassResult
Problem type:classification
Parameters:
An object of class MessinaParameters
5339 features, 38 samples.
Objective type: sensitivity/specificity. Minimum sensitivity: 0.95 Minimum specificity: 0.85
Minimum group fraction: 0
Training fraction: 0.9
Number of bootstraps: 50
Random seed:
Summary of results:
An object of class MessinaFits
166 / 5339 features passed performance requirements (3.11%)
Top features:
Passed Requirements Classifier Type Threshold Value Direction
204719_at TRUE Threshold 6.326002 -1
207502_at TRUE Threshold 8.048459 -1
206784_at TRUE Threshold 10.339348 -1
206134_at TRUE Threshold 15.667098 -1
207003_at TRUE Threshold 10.530530 -1
213921_at TRUE Threshold 4.882769 -1
204259_at TRUE Threshold 2.781876 1
209735_at TRUE Threshold 6.470396 -1
205950_s_at TRUE Threshold 14.161574 -1
206422_at TRUE Threshold 11.371876 -1
Margin
204719_at 7.228815
207502_at 6.366498
206784_at 6.205261
206134_at 6.043961
207003_at 6.013640
213921_at 5.706219
204259_at 4.928655
209735_at 4.927077
205950_s_at 4.368962
206422_at 3.950427
> plot(fit)
Warning message:
Stacking not well defined when ymin != 0
>
>
>
>
>
> dev.off()
null device
1
>
|