Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Plot figures for PCA/PLS-DA loadingsDescriptionPlot figure for PCA/PLS-DA loadings UsageplotLoading(object, out.tol = 0.9, label = 0, fig = "loading.png") Arguments
Valuenone Author(s)Bo Wen wenbo@genomics.cn See Also
Examples
para <- new("metaXpara")
pfile <- system.file("extdata/MTBLS79.txt",package = "metaX")
sfile <- system.file("extdata/MTBLS79_sampleList.txt",package = "metaX")
rawPeaks(para) <- read.delim(pfile,check.names = FALSE)
sampleListFile(para) <- sfile
para <- reSetPeaksData(para)
para <- missingValueImpute(para)
para <- transformation(para,valueID = "value")
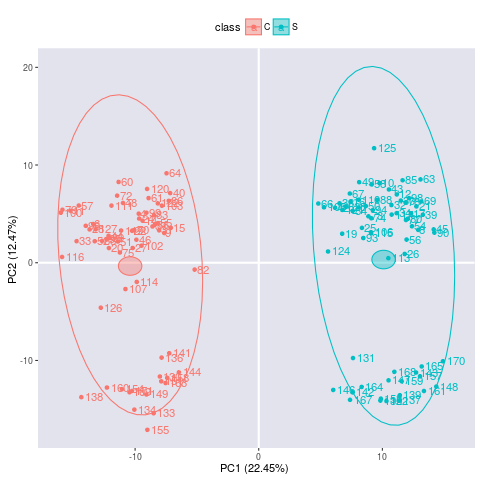
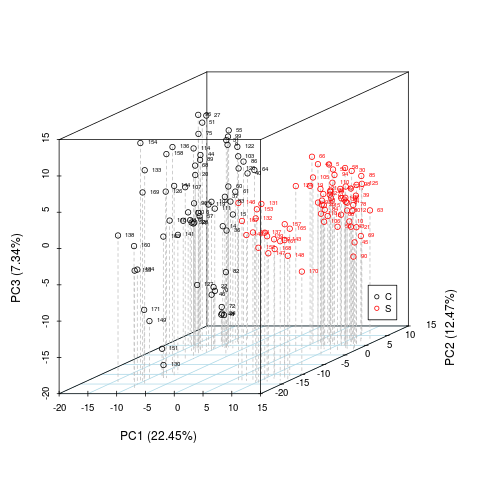
res <- metaX::plotPCA(para,valueID="value",scale="uv",center=TRUE)
plotLoading(res$pca,fig="loading.png")
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(metaX)
Loading required package: VennDiagram
Loading required package: grid
Loading required package: futile.logger
Loading required package: pROC
Type 'citation("pROC")' for a citation.
Attaching package: 'pROC'
The following objects are masked from 'package:stats':
cov, smooth, var
Loading required package: SSPA
Loading required package: qvalue
Loading required package: lattice
Loading required package: limma
Attaching package: 'metaX'
The following object is masked from 'package:xcms':
plotQC
The following objects are masked from 'package:BiocGenerics':
normalize, plotPCA
Warning message:
Package 'metaX' is deprecated and will be removed from Bioconductor
version 3.3
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/metaX/plotLoading.Rd_%03d_medium.png", width=480, height=480)
> ### Name: plotLoading
> ### Title: Plot figures for PCA/PLS-DA loadings
> ### Aliases: plotLoading
>
> ### ** Examples
>
> para <- new("metaXpara")
> pfile <- system.file("extdata/MTBLS79.txt",package = "metaX")
> sfile <- system.file("extdata/MTBLS79_sampleList.txt",package = "metaX")
> rawPeaks(para) <- read.delim(pfile,check.names = FALSE)
> sampleListFile(para) <- sfile
> para <- reSetPeaksData(para)
> para <- missingValueImpute(para)
missingValueImpute: value
Wed Jul 6 21:53:48 2016 Missing value imputation for 'value'
Missing value in total: 3940
Missing value in QC sample: 678
Missing value in non-QC sample: 3262
Wed Jul 6 21:53:48 2016 The ratio of missing value: 4.5814%
<=0: 0
Missing value in total after missing value inputation: 0
<=0 value in total after missing value inputation: 0
> para <- transformation(para,valueID = "value")
> res <- metaX::plotPCA(para,valueID="value",scale="uv",center=TRUE)
plot PCA for value 'value'
> plotLoading(res$pca,fig="loading.png")
pdf
3
>
>
>
>
>
> dev.off()
null device
1
>
|