Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
P-values and test-statistics from the Hedenfalk et al. (2001) gene expression datasetDescriptionThe data from the breast cancer gene expression study of Hedenfalk et al. (2001) were obtained and analyzed. A comparison was made between 3,226 genes of two mutation types, BRCA1 (7 arrays) and BRCA2 (8 arrays). The data included here are p-values, test-statistics, and permutation null test-statistics obtained from a two-sample t-test analysis on a set of 3170 genes, as described in Storey and Tibshirani (2003). Usagedata(hedenfalk) ValueA list called
ReferencesHedenfalk I et al. (2001). Gene expression profiles in hereditary breast cancer. New England Journal of Medicine, 344: 539-548. Storey JD and Tibshirani R. (2003). Statistical significance for genome-wide
studies. Proceedings of the National Academy of Sciences, 100: 9440-9445. See Also
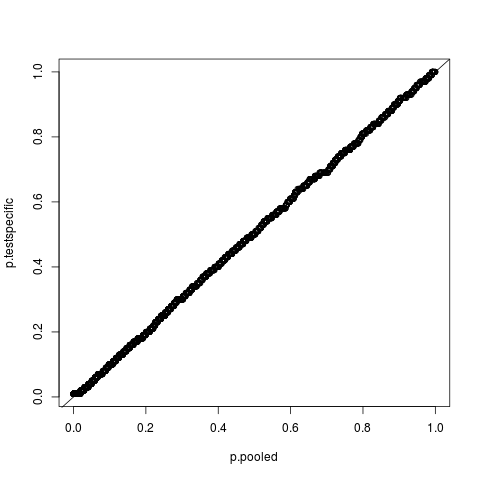
Examples# import data data(hedenfalk) stat <- hedenfalk$stat stat0 <- hedenfalk$stat0 #vector from null distribution p.pooled <- empPvals(stat=stat, stat0=stat0) p.testspecific <- empPvals(stat=stat, stat0=stat0, pool=FALSE) #compare pooled to test-specific p-values qqplot(p.pooled, p.testspecific); abline(0,1) # calculate q-values and view results qobj <- qvalue(p.pooled) summary(qobj) hist(qobj) plot(qobj) Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(qvalue)
> png(filename="/home/ddbj/snapshot/RGM3/R_BC/result/qvalue/hedenfalk.Rd_%03d_medium.png", width=480, height=480)
> ### Name: hedenfalk
> ### Title: P-values and test-statistics from the Hedenfalk et al. (2001)
> ### gene expression dataset
> ### Aliases: hedenfalk
> ### Keywords: dataset, hedenfalk
>
> ### ** Examples
>
> # import data
> data(hedenfalk)
> stat <- hedenfalk$stat
> stat0 <- hedenfalk$stat0 #vector from null distribution
>
> p.pooled <- empPvals(stat=stat, stat0=stat0)
> p.testspecific <- empPvals(stat=stat, stat0=stat0, pool=FALSE)
>
> #compare pooled to test-specific p-values
> qqplot(p.pooled, p.testspecific); abline(0,1)
>
> # calculate q-values and view results
> qobj <- qvalue(p.pooled)
> summary(qobj)
Call:
qvalue(p = p.pooled)
pi0: 0.669926
Cumulative number of significant calls:
<1e-04 <0.001 <0.01 <0.025 <0.05 <0.1 <1
p-value 15 76 265 424 605 868 3170
q-value 0 0 1 73 162 319 3170
local FDR 0 0 3 30 85 167 2241
> hist(qobj)
> plot(qobj)
>
>
>
>
>
>
> dev.off()
null device
1
>
|