Supported by Dr. Osamu Ogasawara and  providing providing  . . |
|
Last data update: 2014.03.03 |
Demand for Medical Care in NMES 1988DescriptionCross-section data originating from the US National Medical Expenditure Survey (NMES) conducted in 1987 and 1988. The NMES is based upon a representative, national probability sample of the civilian non-institutionalized population and individuals admitted to long-term care facilities during 1987. The data are a subsample of individuals ages 66 and over all of whom are covered by Medicare (a public insurance program providing substantial protection against health-care costs). Usagedata("NMES1988")
FormatA data frame containing 4,406 observations on 19 variables.
SourceJournal of Applied Econometrics Data Archive for Deb and Trivedi (1997). http://qed.econ.queensu.ca/jae/1997-v12.3/deb-trivedi/ ReferencesCameron, A.C. and Trivedi, P.K. (1998). Regression Analysis of Count Data. Cambridge: Cambridge University Press. Deb, P., and Trivedi, P.K. (1997). Demand for Medical Care by the Elderly: A Finite Mixture Approach. Journal of Applied Econometrics, 12, 313–336. Zeileis, A., Kleiber, C., and Jackman, S. (2008). Regression Models for Count Data in R. Journal of Statistical Software, 27(8). URL http://www.jstatsoft.org/v27/i08/. See Also
Examples
## packages
library("MASS")
library("pscl")
## select variables for analysis
data("NMES1988")
nmes <- NMES1988[, c(1, 6:8, 13, 15, 18)]
## dependent variable
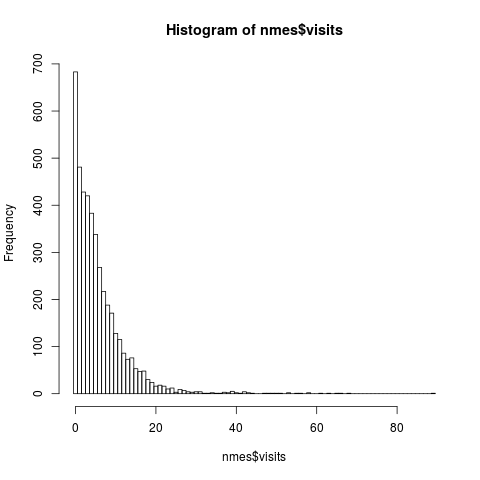
hist(nmes$visits, breaks = 0:(max(nmes$visits)+1) - 0.5)
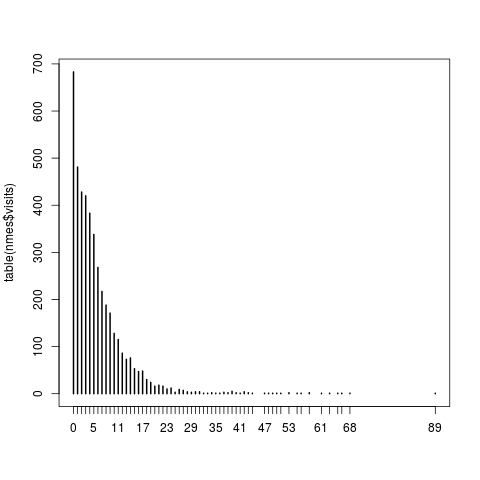
plot(table(nmes$visits))
## convenience transformations for exploratory graphics
clog <- function(x) log(x + 0.5)
cfac <- function(x, breaks = NULL) {
if(is.null(breaks)) breaks <- unique(quantile(x, 0:10/10))
x <- cut(x, breaks, include.lowest = TRUE, right = FALSE)
levels(x) <- paste(breaks[-length(breaks)], ifelse(diff(breaks) > 1,
c(paste("-", breaks[-c(1, length(breaks))] - 1, sep = ""), "+"), ""), sep = "")
return(x)
}
## bivariate visualization
par(mfrow = c(3, 2))
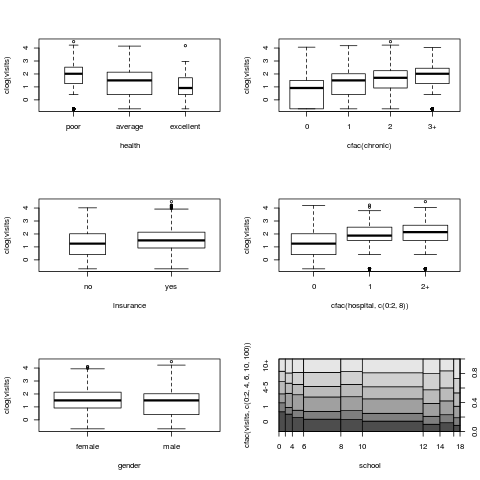
plot(clog(visits) ~ health, data = nmes, varwidth = TRUE)
plot(clog(visits) ~ cfac(chronic), data = nmes)
plot(clog(visits) ~ insurance, data = nmes, varwidth = TRUE)
plot(clog(visits) ~ cfac(hospital, c(0:2, 8)), data = nmes)
plot(clog(visits) ~ gender, data = nmes, varwidth = TRUE)
plot(cfac(visits, c(0:2, 4, 6, 10, 100)) ~ school, data = nmes, breaks = 9)
par(mfrow = c(1, 1))
## Poisson regression
nmes_pois <- glm(visits ~ ., data = nmes, family = poisson)
summary(nmes_pois)
## LM test for overdispersion
dispersiontest(nmes_pois)
dispersiontest(nmes_pois, trafo = 2)
## sandwich covariance matrix
coeftest(nmes_pois, vcov = sandwich)
## quasipoisson model
nmes_qpois <- glm(visits ~ ., data = nmes, family = quasipoisson)
## NegBin regression
nmes_nb <- glm.nb(visits ~ ., data = nmes)
## hurdle regression
nmes_hurdle <- hurdle(visits ~ . | hospital + chronic + insurance + school + gender,
data = nmes, dist = "negbin")
## zero-inflated regression model
nmes_zinb <- zeroinfl(visits ~ . | hospital + chronic + insurance + school + gender,
data = nmes, dist = "negbin")
## compare estimated coefficients
fm <- list("ML-Pois" = nmes_pois, "Quasi-Pois" = nmes_qpois, "NB" = nmes_nb,
"Hurdle-NB" = nmes_hurdle, "ZINB" = nmes_zinb)
round(sapply(fm, function(x) coef(x)[1:8]), digits = 3)
## associated standard errors
round(cbind("ML-Pois" = sqrt(diag(vcov(nmes_pois))),
"Adj-Pois" = sqrt(diag(sandwich(nmes_pois))),
sapply(fm[-1], function(x) sqrt(diag(vcov(x)))[1:8])),
digits = 3)
## log-likelihoods and number of estimated parameters
rbind(logLik = sapply(fm, function(x) round(logLik(x), digits = 0)),
Df = sapply(fm, function(x) attr(logLik(x), "df")))
## predicted number of zeros
round(c("Obs" = sum(nmes$visits < 1),
"ML-Pois" = sum(dpois(0, fitted(nmes_pois))),
"Adj-Pois" = NA,
"Quasi-Pois" = NA,
"NB" = sum(dnbinom(0, mu = fitted(nmes_nb), size = nmes_nb$theta)),
"NB-Hurdle" = sum(predict(nmes_hurdle, type = "prob")[,1]),
"ZINB" = sum(predict(nmes_zinb, type = "prob")[,1])))
## coefficients of zero-augmentation models
t(sapply(fm[4:5], function(x) round(x$coefficients$zero, digits = 3)))
Results
R version 3.3.1 (2016-06-21) -- "Bug in Your Hair"
Copyright (C) 2016 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(AER)
Loading required package: car
Loading required package: lmtest
Loading required package: zoo
Attaching package: 'zoo'
The following objects are masked from 'package:base':
as.Date, as.Date.numeric
Loading required package: sandwich
Loading required package: survival
> png(filename="/home/ddbj/snapshot/RGM3/R_CC/result/AER/NMES1988.Rd_%03d_medium.png", width=480, height=480)
> ### Name: NMES1988
> ### Title: Demand for Medical Care in NMES 1988
> ### Aliases: NMES1988
> ### Keywords: datasets
>
> ### ** Examples
>
> ## packages
> library("MASS")
> library("pscl")
Loading required package: lattice
Classes and Methods for R developed in the
Political Science Computational Laboratory
Department of Political Science
Stanford University
Simon Jackman
hurdle and zeroinfl functions by Achim Zeileis
>
> ## select variables for analysis
> data("NMES1988")
> nmes <- NMES1988[, c(1, 6:8, 13, 15, 18)]
>
> ## dependent variable
> hist(nmes$visits, breaks = 0:(max(nmes$visits)+1) - 0.5)
> plot(table(nmes$visits))
>
> ## convenience transformations for exploratory graphics
> clog <- function(x) log(x + 0.5)
> cfac <- function(x, breaks = NULL) {
+ if(is.null(breaks)) breaks <- unique(quantile(x, 0:10/10))
+ x <- cut(x, breaks, include.lowest = TRUE, right = FALSE)
+ levels(x) <- paste(breaks[-length(breaks)], ifelse(diff(breaks) > 1,
+ c(paste("-", breaks[-c(1, length(breaks))] - 1, sep = ""), "+"), ""), sep = "")
+ return(x)
+ }
>
> ## bivariate visualization
> par(mfrow = c(3, 2))
> plot(clog(visits) ~ health, data = nmes, varwidth = TRUE)
> plot(clog(visits) ~ cfac(chronic), data = nmes)
> plot(clog(visits) ~ insurance, data = nmes, varwidth = TRUE)
> plot(clog(visits) ~ cfac(hospital, c(0:2, 8)), data = nmes)
> plot(clog(visits) ~ gender, data = nmes, varwidth = TRUE)
> plot(cfac(visits, c(0:2, 4, 6, 10, 100)) ~ school, data = nmes, breaks = 9)
> par(mfrow = c(1, 1))
>
> ## Poisson regression
> nmes_pois <- glm(visits ~ ., data = nmes, family = poisson)
> summary(nmes_pois)
Call:
glm(formula = visits ~ ., family = poisson, data = nmes)
Deviance Residuals:
Min 1Q Median 3Q Max
-8.4055 -1.9962 -0.6737 0.7049 16.3620
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.028874 0.023785 43.258 <2e-16 ***
hospital 0.164797 0.005997 27.478 <2e-16 ***
healthpoor 0.248307 0.017845 13.915 <2e-16 ***
healthexcellent -0.361993 0.030304 -11.945 <2e-16 ***
chronic 0.146639 0.004580 32.020 <2e-16 ***
gendermale -0.112320 0.012945 -8.677 <2e-16 ***
school 0.026143 0.001843 14.182 <2e-16 ***
insuranceyes 0.201687 0.016860 11.963 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 26943 on 4405 degrees of freedom
Residual deviance: 23168 on 4398 degrees of freedom
AIC: 35959
Number of Fisher Scoring iterations: 5
>
> ## LM test for overdispersion
> dispersiontest(nmes_pois)
Overdispersion test
data: nmes_pois
z = 11.509, p-value < 2.2e-16
alternative hypothesis: true dispersion is greater than 1
sample estimates:
dispersion
6.706192
> dispersiontest(nmes_pois, trafo = 2)
Overdispersion test
data: nmes_pois
z = 11.374, p-value < 2.2e-16
alternative hypothesis: true alpha is greater than 0
sample estimates:
alpha
0.8953912
>
> ## sandwich covariance matrix
> coeftest(nmes_pois, vcov = sandwich)
z test of coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.028874 0.064530 15.9442 < 2.2e-16 ***
hospital 0.164797 0.021945 7.5095 5.935e-14 ***
healthpoor 0.248307 0.054022 4.5964 4.298e-06 ***
healthexcellent -0.361993 0.077449 -4.6740 2.954e-06 ***
chronic 0.146639 0.012908 11.3605 < 2.2e-16 ***
gendermale -0.112320 0.035343 -3.1780 0.001483 **
school 0.026143 0.005084 5.1422 2.715e-07 ***
insuranceyes 0.201687 0.043128 4.6765 2.919e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
>
> ## quasipoisson model
> nmes_qpois <- glm(visits ~ ., data = nmes, family = quasipoisson)
>
> ## NegBin regression
> nmes_nb <- glm.nb(visits ~ ., data = nmes)
>
> ## hurdle regression
> nmes_hurdle <- hurdle(visits ~ . | hospital + chronic + insurance + school + gender,
+ data = nmes, dist = "negbin")
>
> ## zero-inflated regression model
> nmes_zinb <- zeroinfl(visits ~ . | hospital + chronic + insurance + school + gender,
+ data = nmes, dist = "negbin")
>
> ## compare estimated coefficients
> fm <- list("ML-Pois" = nmes_pois, "Quasi-Pois" = nmes_qpois, "NB" = nmes_nb,
+ "Hurdle-NB" = nmes_hurdle, "ZINB" = nmes_zinb)
> round(sapply(fm, function(x) coef(x)[1:8]), digits = 3)
ML-Pois Quasi-Pois NB Hurdle-NB ZINB
(Intercept) 1.029 1.029 0.929 1.198 1.194
hospital 0.165 0.165 0.218 0.212 0.201
healthpoor 0.248 0.248 0.305 0.316 0.285
healthexcellent -0.362 -0.362 -0.342 -0.332 -0.319
chronic 0.147 0.147 0.175 0.126 0.129
gendermale -0.112 -0.112 -0.126 -0.068 -0.080
school 0.026 0.026 0.027 0.021 0.021
insuranceyes 0.202 0.202 0.224 0.100 0.126
>
> ## associated standard errors
> round(cbind("ML-Pois" = sqrt(diag(vcov(nmes_pois))),
+ "Adj-Pois" = sqrt(diag(sandwich(nmes_pois))),
+ sapply(fm[-1], function(x) sqrt(diag(vcov(x)))[1:8])),
+ digits = 3)
ML-Pois Adj-Pois Quasi-Pois NB Hurdle-NB ZINB
(Intercept) 0.024 0.065 0.062 0.055 0.059 0.057
hospital 0.006 0.022 0.016 0.020 0.021 0.020
healthpoor 0.018 0.054 0.046 0.049 0.048 0.045
healthexcellent 0.030 0.077 0.078 0.061 0.066 0.060
chronic 0.005 0.013 0.012 0.012 0.012 0.012
gendermale 0.013 0.035 0.034 0.031 0.032 0.031
school 0.002 0.005 0.005 0.004 0.005 0.004
insuranceyes 0.017 0.043 0.044 0.039 0.043 0.042
>
> ## log-likelihoods and number of estimated parameters
> rbind(logLik = sapply(fm, function(x) round(logLik(x), digits = 0)),
+ Df = sapply(fm, function(x) attr(logLik(x), "df")))
ML-Pois Quasi-Pois NB Hurdle-NB ZINB
logLik -17972 NA -12171 -12090 -12091
Df 8 8 9 15 15
>
> ## predicted number of zeros
> round(c("Obs" = sum(nmes$visits < 1),
+ "ML-Pois" = sum(dpois(0, fitted(nmes_pois))),
+ "Adj-Pois" = NA,
+ "Quasi-Pois" = NA,
+ "NB" = sum(dnbinom(0, mu = fitted(nmes_nb), size = nmes_nb$theta)),
+ "NB-Hurdle" = sum(predict(nmes_hurdle, type = "prob")[,1]),
+ "ZINB" = sum(predict(nmes_zinb, type = "prob")[,1])))
Obs ML-Pois Adj-Pois Quasi-Pois NB NB-Hurdle ZINB
683 47 NA NA 608 683 709
>
> ## coefficients of zero-augmentation models
> t(sapply(fm[4:5], function(x) round(x$coefficients$zero, digits = 3)))
(Intercept) hospital chronic insuranceyes school gendermale
Hurdle-NB 0.016 0.318 0.548 0.746 0.057 -0.419
ZINB -0.047 -0.800 -1.248 -1.176 -0.084 0.648
>
>
>
>
>
> dev.off()
null device
1
>
|